You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003003_00582
You are here: Home > Sequence: MGYG000003003_00582
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | CAG-1427 sp900539675 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Coriobacteriia; Coriobacteriales; Eggerthellaceae; CAG-1427; CAG-1427 sp900539675 | |||||||||||
| CAZyme ID | MGYG000003003_00582 | |||||||||||
| CAZy Family | CBM13 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 17782; End: 21390 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM13 | 873 | 1027 | 1.3e-28 | 0.7606382978723404 |
| CBM13 | 327 | 461 | 6.6e-23 | 0.6702127659574468 |
| CBM13 | 732 | 866 | 5.3e-18 | 0.675531914893617 |
| CBM13 | 473 | 564 | 8.7e-18 | 0.46808510638297873 |
| CBM13 | 629 | 730 | 3.2e-16 | 0.4787234042553192 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam14200 | RicinB_lectin_2 | 4.18e-20 | 907 | 999 | 1 | 89 | Ricin-type beta-trefoil lectin domain-like. |
| pfam14200 | RicinB_lectin_2 | 5.93e-20 | 350 | 439 | 1 | 89 | Ricin-type beta-trefoil lectin domain-like. |
| pfam14200 | RicinB_lectin_2 | 5.04e-19 | 671 | 761 | 4 | 89 | Ricin-type beta-trefoil lectin domain-like. |
| cd16913 | YkuD_like | 1.60e-18 | 1089 | 1201 | 1 | 121 | L,D-transpeptidases/carboxypeptidases similar to Bacillus YkuD. Members of the YkuD-like family of proteins are found in a range of bacteria. The best studied member Bacillus YkuD has been shown to act as an L,D-transpeptidase that gives rise to an alternative pathway for peptidoglycan cross-linking. Another member Helicobacter pylori Csd6 functions as an L,D-carboxypeptidase and regulates helical cell shape and motility. The conserved region contains a conserved histidine and cysteine, with the cysteine thought to be an active site residue. |
| NF035929 | lectin_1 | 2.64e-17 | 882 | 1011 | 721 | 836 | lectin. Lectins are important adhesin proteins, which bind carbohydrate structures on host cell surface. The carbohydrate specificity of diverse lectins to a large extent dictates bacteria tissue tropism by mediating specific attachment to unique host sites expressing the corresponding carbohydrate receptor. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QRT49575.1 | 4.05e-111 | 297 | 1066 | 109 | 871 |
| BAK44091.1 | 9.32e-72 | 299 | 1086 | 128 | 906 |
| QRT49576.1 | 5.35e-55 | 461 | 981 | 414 | 928 |
| AMK55176.1 | 2.30e-27 | 308 | 777 | 509 | 975 |
| QOL34941.1 | 3.23e-21 | 297 | 530 | 753 | 985 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4XXT_A | 1.13e-15 | 1086 | 1201 | 139 | 261 | Crystalstructure of Fused Zn-dependent amidase/peptidase/peptodoglycan-binding domain-containing protein from Clostridium acetobutylicum ATCC 824 [Clostridium acetobutylicum ATCC 824] |
| 4OUJ_A | 5.09e-06 | 628 | 913 | 1 | 292 | Crystalstructure of HA33B-Lac [Clostridium botulinum B1 str. Okra],4OUJ_B Crystal structure of HA33B-Lac [Clostridium botulinum B1 str. Okra] |
| 4LO0_A | 8.46e-06 | 630 | 913 | 1 | 289 | ApoHA17-HA33 [Clostridium botulinum],4LO0_B Apo HA17-HA33 [Clostridium botulinum],4LO1_A HA17-HA33-Gal [Clostridium botulinum],4LO1_B HA17-HA33-Gal [Clostridium botulinum],4LO2_A HA17-HA33-Lac [Clostridium botulinum],4LO2_B HA17-HA33-Lac [Clostridium botulinum],4LO3_A HA17-HA33-LacNac [Clostridium botulinum],4LO3_B HA17-HA33-LacNac [Clostridium botulinum],4LO7_B HA70(D3)-HA17-HA33 [Clostridium botulinum],4LO7_D HA70(D3)-HA17-HA33 [Clostridium botulinum],4LO7_F HA70(D3)-HA17-HA33 [Clostridium botulinum],4LO7_H HA70(D3)-HA17-HA33 [Clostridium botulinum],4QD2_C Molecular basis for disruption of E-cadherin adhesion by botulinum neurotoxin A complex [Clostridium botulinum A str. Hall],4QD2_D Molecular basis for disruption of E-cadherin adhesion by botulinum neurotoxin A complex [Clostridium botulinum A str. Hall],4QD2_H Molecular basis for disruption of E-cadherin adhesion by botulinum neurotoxin A complex [Clostridium botulinum A str. Hall],4QD2_I Molecular basis for disruption of E-cadherin adhesion by botulinum neurotoxin A complex [Clostridium botulinum A str. Hall],5BP5_A Crystal structure of HA17-HA33-IPT [Clostridium botulinum],5BP5_B Crystal structure of HA17-HA33-IPT [Clostridium botulinum],5BQU_A Crystal structure of HA17-HA33-Lactulose [Clostridium botulinum],5BQU_B Crystal structure of HA17-HA33-Lactulose [Clostridium botulinum] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
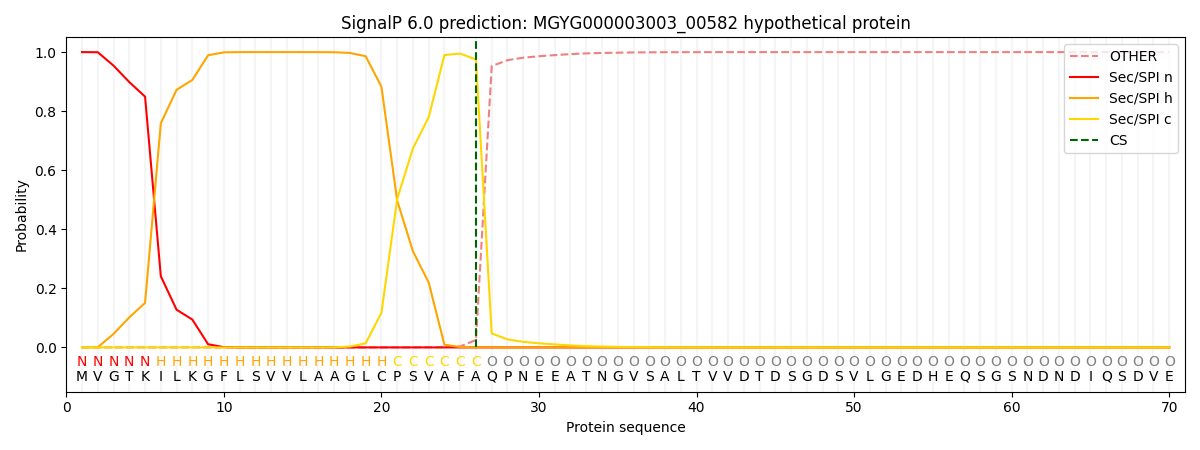
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000298 | 0.998934 | 0.000253 | 0.000172 | 0.000172 | 0.000152 |
