You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003005_01903
You are here: Home > Sequence: MGYG000003005_01903
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
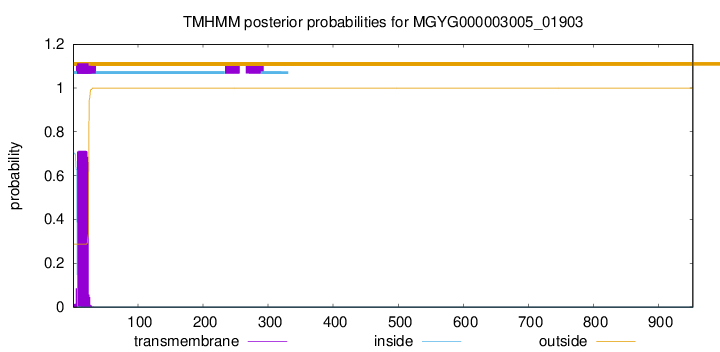
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Ruminococcaceae; Ruminococcus; | |||||||||||
| CAZyme ID | MGYG000003005_01903 | |||||||||||
| CAZy Family | PL1 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 666; End: 3527 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL1 | 211 | 433 | 1.1e-34 | 0.9257425742574258 |
| CBM13 | 557 | 702 | 1.8e-23 | 0.7021276595744681 |
| CBM13 | 711 | 865 | 5.4e-17 | 0.75 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3866 | PelB | 8.03e-29 | 167 | 513 | 30 | 343 | Pectate lyase [Carbohydrate transport and metabolism]. |
| smart00656 | Amb_all | 1.04e-19 | 236 | 433 | 17 | 188 | Amb_all domain. |
| pfam14200 | RicinB_lectin_2 | 3.72e-16 | 642 | 741 | 1 | 89 | Ricin-type beta-trefoil lectin domain-like. |
| pfam14200 | RicinB_lectin_2 | 4.70e-16 | 558 | 636 | 15 | 89 | Ricin-type beta-trefoil lectin domain-like. |
| pfam14200 | RicinB_lectin_2 | 4.60e-15 | 596 | 685 | 5 | 89 | Ricin-type beta-trefoil lectin domain-like. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| CBL17651.1 | 1.91e-181 | 33 | 948 | 34 | 893 |
| CDM70399.1 | 9.74e-166 | 33 | 700 | 31 | 696 |
| AUO18238.1 | 5.34e-146 | 18 | 700 | 19 | 707 |
| CBL16867.1 | 1.02e-140 | 8 | 947 | 10 | 912 |
| ASR46637.1 | 1.14e-82 | 33 | 530 | 35 | 536 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3KRG_A | 3.27e-13 | 267 | 482 | 149 | 374 | ChainA, Pectate lyase [Bacillus subtilis] |
| 5AMV_A | 3.27e-13 | 267 | 482 | 149 | 374 | Structuralinsights into the loss of catalytic competence in pectate lyase at low pH [Bacillus subtilis],5X2I_A Polygalacturonate Lyase by Fusing with a Self-assembling Amphipathic Peptide [Bacillus subtilis subsp. subtilis str. 168] |
| 1BN8_A | 3.70e-13 | 267 | 482 | 170 | 395 | BacillusSubtilis Pectate Lyase [Bacillus subtilis] |
| 2BSP_A | 8.66e-13 | 267 | 482 | 170 | 395 | ChainA, PROTEIN (PECTATE LYASE) [Bacillus subtilis] |
| 2NZM_A | 1.81e-12 | 267 | 482 | 149 | 374 | ChainA, Pectate lyase [Bacillus subtilis],2O04_A Chain A, Pectate lyase [Bacillus subtilis],2O0V_A Chain A, Pectate lyase [Bacillus subtilis],2O0W_A Chain A, Pectate lyase [Bacillus subtilis],2O17_A Chain A, Pectate lyase [Bacillus subtilis],2O1D_A Chain A, Pectate lyase [Bacillus subtilis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P94449 | 3.99e-22 | 169 | 511 | 34 | 336 | Pectin lyase OS=Bacillus subtilis OX=1423 GN=pelB PE=1 SV=1 |
| O34819 | 5.82e-21 | 169 | 511 | 34 | 336 | Pectin lyase OS=Bacillus subtilis (strain 168) OX=224308 GN=pelB PE=3 SV=1 |
| P24112 | 1.22e-16 | 167 | 492 | 7 | 291 | Pectin lyase OS=Pectobacterium carotovorum OX=554 GN=pnl PE=1 SV=1 |
| P27027 | 3.09e-15 | 186 | 511 | 18 | 303 | Pectin lyase OS=Pseudomonas marginalis OX=298 GN=pnl PE=1 SV=2 |
| P39116 | 2.03e-12 | 267 | 482 | 170 | 395 | Pectate lyase OS=Bacillus subtilis (strain 168) OX=224308 GN=pel PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
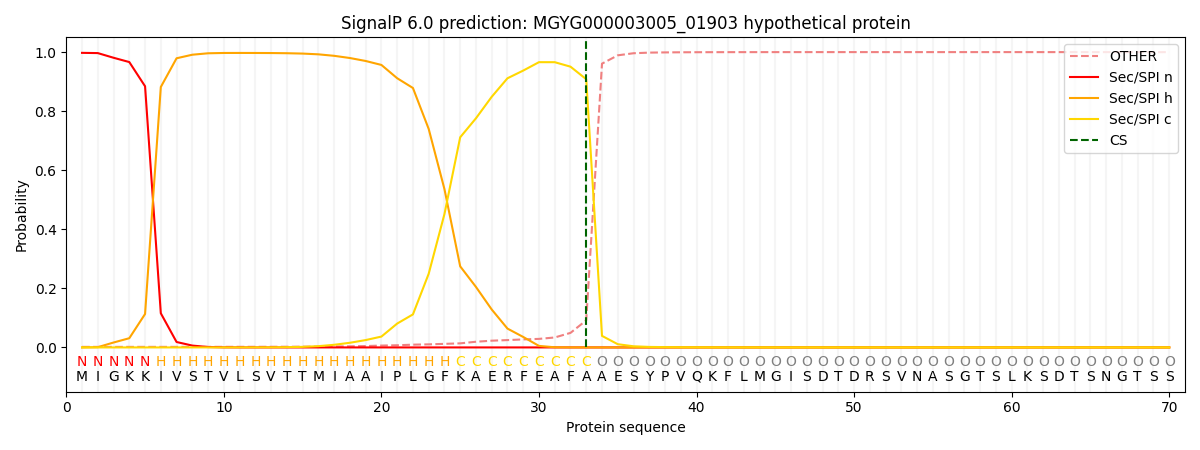
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.003531 | 0.994820 | 0.000974 | 0.000251 | 0.000204 | 0.000187 |