You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003013_00245
You are here: Home > Sequence: MGYG000003013_00245
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Oribacterium sinus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Oribacterium; Oribacterium sinus | |||||||||||
| CAZyme ID | MGYG000003013_00245 | |||||||||||
| CAZy Family | GH73 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 35422; End: 36981 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG4193 | LytD | 5.25e-16 | 335 | 446 | 144 | 245 | Beta- N-acetylglucosaminidase [Carbohydrate transport and metabolism]. |
| pfam08239 | SH3_3 | 1.63e-06 | 23 | 77 | 3 | 54 | Bacterial SH3 domain. |
| smart00287 | SH3b | 6.52e-04 | 16 | 77 | 4 | 62 | Bacterial SH3 domain homologues. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QRV21758.1 | 1.60e-130 | 12 | 465 | 37 | 526 |
| ADL03932.1 | 1.60e-130 | 12 | 465 | 37 | 526 |
| CBK77705.1 | 7.67e-123 | 1 | 465 | 27 | 548 |
| ASN95383.1 | 5.04e-122 | 16 | 465 | 42 | 602 |
| QRP39925.1 | 5.04e-122 | 16 | 465 | 42 | 602 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6FXO_A | 7.85e-15 | 300 | 446 | 97 | 244 | ChainA, Bifunctional autolysin [Staphylococcus aureus subsp. aureus Mu50] |
| 4PI7_A | 8.47e-12 | 297 | 431 | 86 | 214 | ChainA, Autolysin E [Staphylococcus aureus subsp. aureus Mu50],4PI9_A Chain A, Autolysin E [Staphylococcus aureus subsp. aureus Mu50],4PIA_A Chain A, Autolysin E [Staphylococcus aureus subsp. aureus Mu50] |
| 4PI8_A | 5.15e-11 | 297 | 431 | 86 | 214 | ChainA, Autolysin E [Staphylococcus aureus subsp. aureus Mu50] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q8CPQ1 | 4.88e-14 | 300 | 446 | 1188 | 1335 | Bifunctional autolysin OS=Staphylococcus epidermidis (strain ATCC 12228 / FDA PCI 1200) OX=176280 GN=atl PE=3 SV=1 |
| O33635 | 1.13e-13 | 300 | 446 | 1188 | 1335 | Bifunctional autolysin OS=Staphylococcus epidermidis OX=1282 GN=atl PE=1 SV=1 |
| Q5HQB9 | 1.13e-13 | 300 | 446 | 1188 | 1335 | Bifunctional autolysin OS=Staphylococcus epidermidis (strain ATCC 35984 / RP62A) OX=176279 GN=atl PE=3 SV=1 |
| Q931U5 | 5.86e-13 | 300 | 446 | 1101 | 1248 | Bifunctional autolysin OS=Staphylococcus aureus (strain Mu50 / ATCC 700699) OX=158878 GN=atl PE=1 SV=2 |
| Q99V41 | 5.86e-13 | 300 | 446 | 1101 | 1248 | Bifunctional autolysin OS=Staphylococcus aureus (strain N315) OX=158879 GN=atl PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
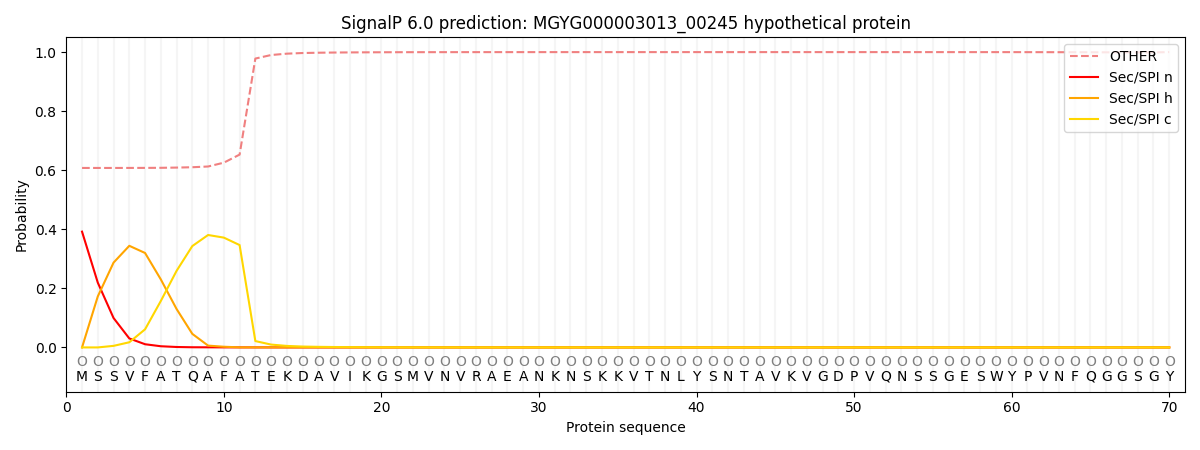
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.614605 | 0.384235 | 0.000297 | 0.000496 | 0.000174 | 0.000180 |
