You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003016_00515
You are here: Home > Sequence: MGYG000003016_00515
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Salinicola tamaricis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Proteobacteria; Gammaproteobacteria; Pseudomonadales; Halomonadaceae; Salinicola; Salinicola tamaricis | |||||||||||
| CAZyme ID | MGYG000003016_00515 | |||||||||||
| CAZy Family | GT83 | |||||||||||
| CAZyme Description | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 9526; End: 11154 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT83 | 22 | 496 | 1.6e-79 | 0.9148148148148149 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG1807 | ArnT | 1.33e-21 | 23 | 321 | 12 | 320 | 4-amino-4-deoxy-L-arabinose transferase or related glycosyltransferase of PMT family [Cell wall/membrane/envelope biogenesis]. |
| pfam13231 | PMT_2 | 7.84e-10 | 74 | 199 | 1 | 127 | Dolichyl-phosphate-mannose-protein mannosyltransferase. This family contains members that are not captured by pfam02366. |
| PRK13279 | arnT | 1.73e-04 | 17 | 317 | 3 | 313 | lipid IV(A) 4-amino-4-deoxy-L-arabinosyltransferase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QXE90238.1 | 1.65e-88 | 23 | 497 | 14 | 496 |
| QXM07636.1 | 1.65e-88 | 23 | 497 | 14 | 496 |
| BBL59367.1 | 5.77e-88 | 25 | 497 | 13 | 497 |
| QWV93130.1 | 6.46e-88 | 23 | 500 | 14 | 499 |
| QXE86324.1 | 3.54e-87 | 23 | 497 | 14 | 496 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5EZM_A | 6.46e-16 | 39 | 338 | 52 | 368 | CrystalStructure of ArnT from Cupriavidus metallidurans in the apo state [Cupriavidus metallidurans CH34],5F15_A Crystal Structure of ArnT from Cupriavidus metallidurans bound to Undecaprenyl phosphate [Cupriavidus metallidurans CH34] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| A6V1N8 | 4.97e-23 | 45 | 417 | 31 | 417 | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase OS=Pseudomonas aeruginosa (strain PA7) OX=381754 GN=arnT PE=3 SV=1 |
| Q02R27 | 2.85e-21 | 45 | 417 | 31 | 417 | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase OS=Pseudomonas aeruginosa (strain UCBPP-PA14) OX=208963 GN=arnT PE=3 SV=1 |
| Q9HY61 | 3.80e-21 | 45 | 417 | 31 | 417 | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase OS=Pseudomonas aeruginosa (strain ATCC 15692 / DSM 22644 / CIP 104116 / JCM 14847 / LMG 12228 / 1C / PRS 101 / PAO1) OX=208964 GN=arnT PE=3 SV=1 |
| B7VBN0 | 1.20e-20 | 45 | 417 | 31 | 417 | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase OS=Pseudomonas aeruginosa (strain LESB58) OX=557722 GN=arnT PE=3 SV=1 |
| Q48HY9 | 2.66e-20 | 17 | 363 | 3 | 361 | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase OS=Pseudomonas savastanoi pv. phaseolicola (strain 1448A / Race 6) OX=264730 GN=arnT PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000024 | 0.000021 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |
TMHMM Annotations download full data without filtering help
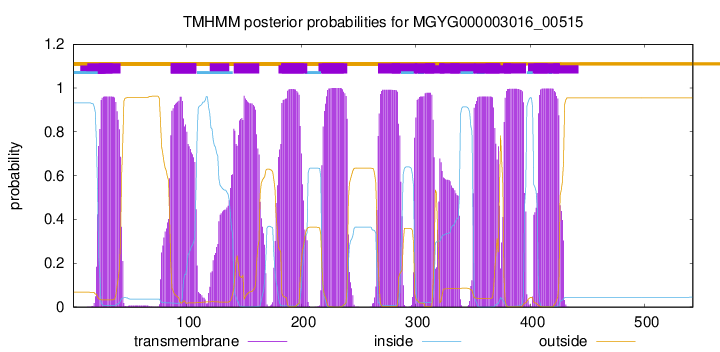
| start | end |
|---|---|
| 23 | 42 |
| 86 | 108 |
| 141 | 163 |
| 183 | 205 |
| 218 | 240 |
| 267 | 286 |
| 299 | 316 |
| 321 | 338 |
| 351 | 373 |
| 377 | 396 |
| 403 | 425 |
