You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003018_00202
You are here: Home > Sequence: MGYG000003018_00202
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Alistipes_A sp900549685 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Rikenellaceae; Alistipes_A; Alistipes_A sp900549685 | |||||||||||
| CAZyme ID | MGYG000003018_00202 | |||||||||||
| CAZy Family | GH109 | |||||||||||
| CAZyme Description | Inositol 2-dehydrogenase/D-chiro-inositol 3-dehydrogenase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 24505; End: 25860 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH109 | 64 | 251 | 1.9e-20 | 0.45614035087719296 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG0673 | MviM | 1.57e-36 | 63 | 419 | 1 | 342 | Predicted dehydrogenase [General function prediction only]. |
| pfam01408 | GFO_IDH_MocA | 1.39e-12 | 66 | 182 | 1 | 106 | Oxidoreductase family, NAD-binding Rossmann fold. This family of enzymes utilize NADP or NAD. This family is called the GFO/IDH/MOCA family in swiss-prot. |
| pfam03435 | Sacchrp_dh_NADP | 2.40e-04 | 70 | 164 | 3 | 94 | Saccharopine dehydrogenase NADP binding domain. This family contains the NADP binding domain of saccharopine dehydrogenase. In some organisms this enzyme is found as a bifunctional polypeptide with lysine ketoglutarate reductase. The saccharopine dehydrogenase can also function as a saccharopine reductase. |
| cd19930 | REC_DesR-like | 0.009 | 126 | 169 | 58 | 101 | phosphoacceptor receiver (REC) domain of DesR and similar proteins. This group is composed of Bacillus subtilis DesR, Streptococcus pneumoniae response regulator spr1814, and similar proteins, all containing an N-terminal REC domain and a C-terminal LuxR family helix-turn-helix (HTH) DNA-binding output domain. DesR is a response regulator that, together with its cognate sensor kinase DesK, comprises a two-component regulatory system that controls membrane fluidity. Phosphorylation of the REC domain of DesR is allosterically coupled to two distinct exposed surfaces of the protein, controlling noncanonical dimerization/tetramerization, cooperative activation, and DesK binding. REC domains function as phosphorylation-mediated switches within response regulators, but some also transfer phosphoryl groups in multistep phosphorelays. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QGA23801.1 | 3.26e-310 | 1 | 451 | 1 | 453 |
| BCG53919.1 | 2.83e-253 | 1 | 451 | 1 | 459 |
| BBK93797.1 | 1.74e-242 | 1 | 448 | 1 | 454 |
| QIX65768.1 | 1.74e-242 | 1 | 448 | 1 | 454 |
| QUT96603.1 | 1.74e-242 | 1 | 448 | 1 | 454 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3CEA_A | 1.53e-14 | 64 | 336 | 7 | 243 | ChainA, Myo-inositol 2-dehydrogenase [Lactiplantibacillus plantarum WCFS1],3CEA_B Chain B, Myo-inositol 2-dehydrogenase [Lactiplantibacillus plantarum WCFS1],3CEA_C Chain C, Myo-inositol 2-dehydrogenase [Lactiplantibacillus plantarum WCFS1],3CEA_D Chain D, Myo-inositol 2-dehydrogenase [Lactiplantibacillus plantarum WCFS1] |
| 3EC7_A | 5.67e-09 | 63 | 316 | 21 | 243 | CrystalStructure of Putative Dehydrogenase from Salmonella typhimurium LT2 [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2],3EC7_B Crystal Structure of Putative Dehydrogenase from Salmonella typhimurium LT2 [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2],3EC7_C Crystal Structure of Putative Dehydrogenase from Salmonella typhimurium LT2 [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2],3EC7_D Crystal Structure of Putative Dehydrogenase from Salmonella typhimurium LT2 [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2],3EC7_E Crystal Structure of Putative Dehydrogenase from Salmonella typhimurium LT2 [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2],3EC7_F Crystal Structure of Putative Dehydrogenase from Salmonella typhimurium LT2 [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2],3EC7_G Crystal Structure of Putative Dehydrogenase from Salmonella typhimurium LT2 [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2],3EC7_H Crystal Structure of Putative Dehydrogenase from Salmonella typhimurium LT2 [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2] |
| 4N54_A | 4.12e-08 | 64 | 410 | 13 | 342 | ChainA, Inositol dehydrogenase [Lacticaseibacillus casei BL23],4N54_B Chain B, Inositol dehydrogenase [Lacticaseibacillus casei BL23],4N54_C Chain C, Inositol dehydrogenase [Lacticaseibacillus casei BL23],4N54_D Chain D, Inositol dehydrogenase [Lacticaseibacillus casei BL23] |
| 4MKX_A | 4.17e-08 | 64 | 410 | 16 | 345 | ChainA, Inositol dehydrogenase [Lacticaseibacillus casei BL23],4MKZ_A Chain A, Inositol dehydrogenase [Lacticaseibacillus casei BL23] |
| 3MOI_A | 5.94e-07 | 139 | 239 | 65 | 161 | Thecrystal structure of the putative dehydrogenase from Bordetella bronchiseptica RB50 [Bordetella bronchiseptica] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q54728 | 1.85e-09 | 66 | 312 | 2 | 221 | Uncharacterized oxidoreductase SP_1686 OS=Streptococcus pneumoniae serotype 4 (strain ATCC BAA-334 / TIGR4) OX=170187 GN=SP_1686 PE=3 SV=2 |
| Q9RK81 | 1.91e-09 | 1 | 297 | 1 | 269 | Glycosyl hydrolase family 109 protein OS=Streptomyces coelicolor (strain ATCC BAA-471 / A3(2) / M145) OX=100226 GN=SCO0529 PE=3 SV=1 |
| A9N564 | 1.19e-08 | 66 | 316 | 3 | 222 | Inositol 2-dehydrogenase OS=Salmonella paratyphi B (strain ATCC BAA-1250 / SPB7) OX=1016998 GN=iolG PE=3 SV=1 |
| Q8ZK57 | 1.19e-08 | 66 | 316 | 3 | 222 | Inositol 2-dehydrogenase OS=Salmonella typhimurium (strain LT2 / SGSC1412 / ATCC 700720) OX=99287 GN=iolG PE=1 SV=1 |
| B5F3F4 | 1.19e-08 | 66 | 316 | 3 | 222 | Inositol 2-dehydrogenase OS=Salmonella agona (strain SL483) OX=454166 GN=iolG PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as TATLIPO
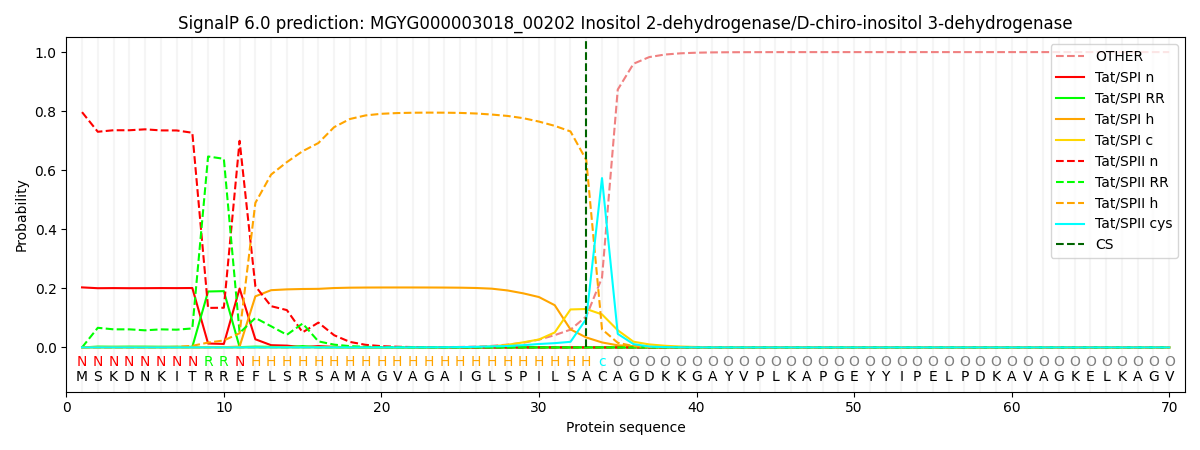
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000001 | 0.000036 | 0.203279 | 0.796688 | 0.000000 |

