You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003035_00061
You are here: Home > Sequence: MGYG000003035_00061
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | CAG-485 sp900542185 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Muribaculaceae; CAG-485; CAG-485 sp900542185 | |||||||||||
| CAZyme ID | MGYG000003035_00061 | |||||||||||
| CAZy Family | GH59 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 87440; End: 89554 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH59 | 30 | 672 | 2.8e-145 | 0.9904912836767037 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam02057 | Glyco_hydro_59 | 4.82e-92 | 35 | 332 | 1 | 293 | Glycosyl hydrolase family 59. |
| pfam17387 | Glyco_hydro_59M | 5.91e-07 | 348 | 435 | 8 | 116 | Glycosyl hydrolase family 59 central domain. |
| pfam06439 | DUF1080 | 1.65e-04 | 500 | 672 | 33 | 181 | Domain of Unknown Function (DUF1080). This family has structural similarity to an endo-1,3-1,4-beta glucanase belonging to glycoside hydrolase family 16. However, the structure surrounding the active site differs from that of the endo-1,3-1,4-beta glucanase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QIH32702.1 | 0.0 | 9 | 693 | 13 | 698 |
| ADY35987.1 | 0.0 | 22 | 693 | 31 | 703 |
| SDU27775.1 | 8.23e-293 | 20 | 693 | 45 | 718 |
| AYV47626.1 | 3.31e-254 | 15 | 673 | 23 | 701 |
| AYD49420.1 | 4.09e-254 | 11 | 693 | 1 | 663 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4CCC_A | 4.70e-85 | 28 | 672 | 18 | 650 | StructureOf Mouse Galactocerebrosidase With 4nbdg: Enzyme-substrate Complex [Mus musculus],4CCD_A Structure Of Mouse Galactocerebrosidase With D-galactal: Enzyme-intermediate Complex [Mus musculus],4CCE_A Structure Of Mouse Galactocerebrosidase With Galactose: Enzyme-product Complex [Mus musculus],4UFH_A Mouse Galactocerebrosidase complexed with iso-galacto-fagomine IGF [Mus musculus],4UFI_A Mouse Galactocerebrosidase complexed with aza-galacto-fagomine AGF [Mus musculus],4UFJ_A Mouse Galactocerebrosidase complexed with iso-galacto-fagomine lactam IGL [Mus musculus],4UFK_A Mouse Galactocerebrosidase complexed with dideoxy-imino-lyxitol DIL [Mus musculus],4UFL_A Mouse Galactocerebrosidase complexed with deoxy-galacto-noeurostegine DGN [Mus musculus],4UFM_A Mouse Galactocerebrosidase complexed with 1-deoxy-galacto-nojirimycin DGJ [Mus musculus],5NXB_A Mouse galactocerebrosidase in complex with saposin A [Mus musculus],5NXB_B Mouse galactocerebrosidase in complex with saposin A [Mus musculus],6Y6S_A Chain A, Galactocerebrosidase [Mus musculus],6Y6T_A Chain A, Galactocerebrosidase [Mus musculus] |
| 3ZR5_A | 4.92e-85 | 28 | 672 | 20 | 652 | STRUCTUREOF GALACTOCEREBROSIDASE FROM MOUSE [Mus musculus],3ZR6_A STRUCTURE OF GALACTOCEREBROSIDASE FROM MOUSE IN COMPLEX WITH GALACTOSE [Mus musculus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q498K0 | 7.46e-96 | 32 | 635 | 44 | 637 | Galactocerebrosidase OS=Xenopus laevis OX=8355 GN=galc PE=2 SV=2 |
| Q0VA39 | 2.93e-95 | 32 | 635 | 44 | 638 | Galactocerebrosidase OS=Xenopus tropicalis OX=8364 GN=galc PE=2 SV=1 |
| P54803 | 1.13e-90 | 11 | 634 | 31 | 646 | Galactocerebrosidase OS=Homo sapiens OX=9606 GN=GALC PE=1 SV=3 |
| Q5SNX7 | 2.39e-90 | 32 | 672 | 30 | 657 | Galactocerebrosidase OS=Danio rerio OX=7955 GN=galc PE=2 SV=1 |
| P54804 | 4.46e-88 | 28 | 639 | 32 | 634 | Galactocerebrosidase OS=Canis lupus familiaris OX=9615 GN=GALC PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
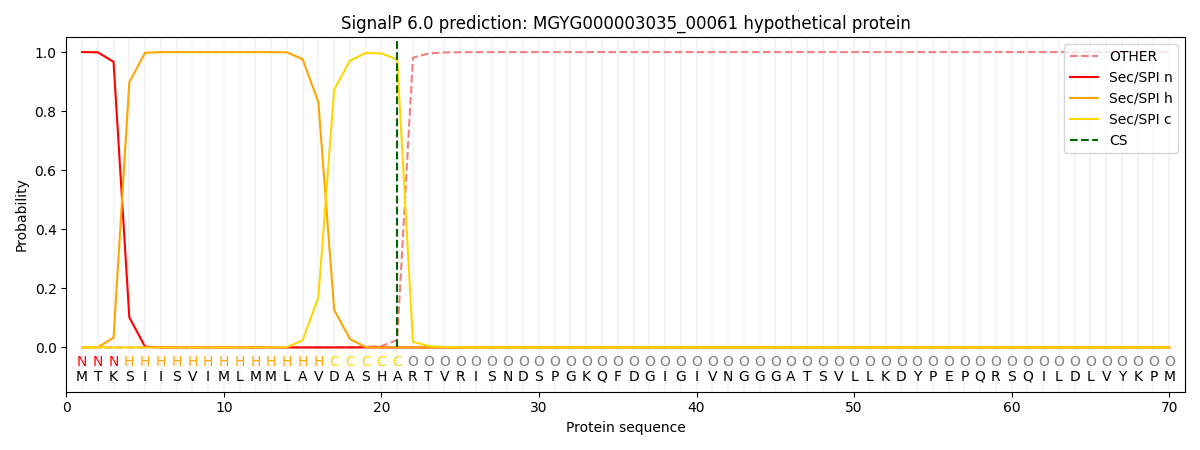
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000257 | 0.999061 | 0.000168 | 0.000170 | 0.000168 | 0.000151 |
