You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003064_00384
You are here: Home > Sequence: MGYG000003064_00384
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
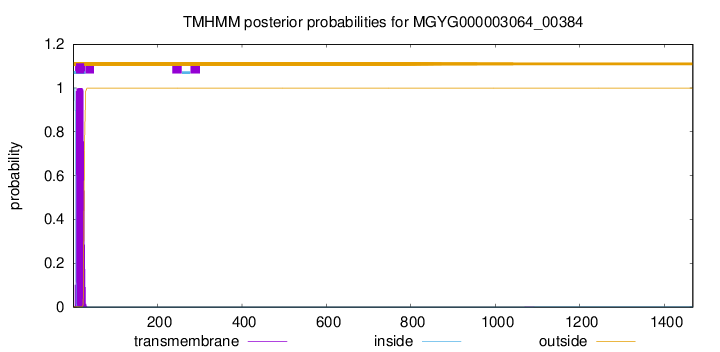
TMHMM annotations
Basic Information help
| Species | Bacteroides graminisolvens | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides graminisolvens | |||||||||||
| CAZyme ID | MGYG000003064_00384 | |||||||||||
| CAZy Family | GH2 | |||||||||||
| CAZyme Description | Beta-galactosidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 483654; End: 488060 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL10 | 1170 | 1458 | 3.3e-93 | 0.9965277777777778 |
| GH2 | 74 | 714 | 4.4e-74 | 0.636968085106383 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| TIGR02474 | pec_lyase | 1.82e-85 | 1170 | 1458 | 1 | 289 | pectate lyase, PelA/Pel-15E family. Members of this family are isozymes of pectate lyase (EC 4.2.2.2), also called polygalacturonic transeliminase and alpha-1,4-D-endopolygalacturonic acid lyase. [Energy metabolism, Biosynthesis and degradation of polysaccharides] |
| pfam09492 | Pec_lyase | 3.74e-84 | 1170 | 1459 | 1 | 289 | Pectic acid lyase. Members of this family are isozymes of pectate lyase (EC:4.2.2.2), also called polygalacturonic transeliminase and alpha-1,4-D-endopolygalacturonic acid lyase. |
| COG3250 | LacZ | 1.12e-34 | 26 | 479 | 7 | 429 | Beta-galactosidase/beta-glucuronidase [Carbohydrate transport and metabolism]. |
| PRK10150 | PRK10150 | 3.91e-22 | 109 | 479 | 68 | 445 | beta-D-glucuronidase; Provisional |
| PRK10340 | ebgA | 3.57e-16 | 112 | 479 | 115 | 472 | cryptic beta-D-galactosidase subunit alpha; Reviewed |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QJR71326.1 | 0.0 | 4 | 1128 | 2 | 1122 |
| QJR62727.1 | 0.0 | 4 | 1128 | 2 | 1122 |
| QJR66986.1 | 0.0 | 4 | 1128 | 2 | 1122 |
| QJR75370.1 | 0.0 | 4 | 1128 | 2 | 1122 |
| AND19553.1 | 0.0 | 4 | 1128 | 2 | 1122 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1R76_A | 1.72e-45 | 1154 | 1468 | 78 | 407 | ChainA, pectate lyase [Niveispirillum irakense] |
| 1GXM_A | 7.01e-32 | 1163 | 1452 | 37 | 316 | Family10 polysaccharide lyase from Cellvibrio cellulosa [Cellvibrio japonicus],1GXM_B Family 10 polysaccharide lyase from Cellvibrio cellulosa [Cellvibrio japonicus],1GXN_A Family 10 polysaccharide lyase from Cellvibrio cellulosa [Cellvibrio japonicus] |
| 1GXO_A | 7.91e-31 | 1163 | 1452 | 37 | 316 | MutantD189A of Family 10 polysaccharide lyase from Cellvibrio cellulosa in complex with trigalaturonic acid [Cellvibrio japonicus] |
| 6ED1_A | 1.66e-29 | 70 | 512 | 45 | 485 | ChainA, Glycosyl hydrolase family 2, sugar binding domain protein [Phocaeicola dorei],6ED1_B Chain B, Glycosyl hydrolase family 2, sugar binding domain protein [Phocaeicola dorei],6ED1_C Chain C, Glycosyl hydrolase family 2, sugar binding domain protein [Phocaeicola dorei],6ED1_D Chain D, Glycosyl hydrolase family 2, sugar binding domain protein [Phocaeicola dorei] |
| 6B6L_A | 2.59e-22 | 76 | 460 | 43 | 414 | Thecrystal structure of glycosyl hydrolase family 2 (GH2) member from Bacteroides cellulosilyticus DSM 14838 [Bacteroides cellulosilyticus DSM 14838],6B6L_B The crystal structure of glycosyl hydrolase family 2 (GH2) member from Bacteroides cellulosilyticus DSM 14838 [Bacteroides cellulosilyticus DSM 14838] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| T2KPJ7 | 6.05e-29 | 108 | 575 | 105 | 542 | Putative beta-glucuronidase OS=Formosa agariphila (strain DSM 15362 / KCTC 12365 / LMG 23005 / KMM 3901 / M-2Alg 35-1) OX=1347342 GN=BN863_21970 PE=2 SV=1 |
| P77989 | 8.42e-23 | 101 | 477 | 51 | 410 | Beta-galactosidase OS=Thermoanaerobacter pseudethanolicus (strain ATCC 33223 / 39E) OX=340099 GN=lacZ PE=3 SV=2 |
| P26257 | 9.44e-22 | 125 | 493 | 73 | 433 | Beta-galactosidase OS=Thermoanaerobacterium thermosulfurigenes OX=33950 GN=lacZ PE=1 SV=1 |
| Q56307 | 2.06e-17 | 110 | 453 | 114 | 443 | Beta-galactosidase OS=Thermotoga maritima (strain ATCC 43589 / DSM 3109 / JCM 10099 / NBRC 100826 / MSB8) OX=243274 GN=lacZ PE=1 SV=2 |
| O52847 | 1.02e-16 | 121 | 507 | 148 | 534 | Beta-galactosidase OS=Priestia megaterium (strain DSM 319 / IMG 1521) OX=592022 GN=bgaM PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
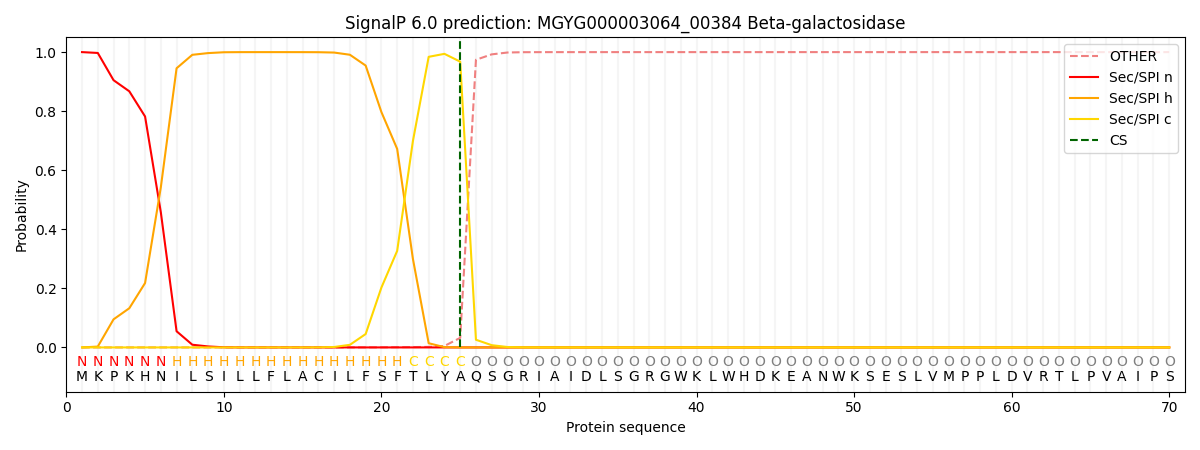
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000583 | 0.998423 | 0.000363 | 0.000203 | 0.000200 | 0.000188 |