You are browsing environment: HUMAN GUT
MGYG000003064_00605
Basic Information
help
Species
Bacteroides graminisolvens
Lineage
Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides graminisolvens
CAZyme ID
MGYG000003064_00605
CAZy Family
CE19
CAZyme Description
hypothetical protein
CAZyme Property
Genome Property
Genome Assembly ID
Genome Size
Genome Type
Country
Continent
MGYG000003064
3410784
MAG
United States
North America
Gene Location
Start: 151018;
End: 152994
Strand: +
No EC number prediction in MGYG000003064_00605.
Family
Start
End
Evalue
family coverage
CE19
119
448
1.4e-138
0.9879518072289156
Cdd ID
Domain
E-Value
qStart
qEnd
sStart
sEnd
Domain Description
pfam12875
DUF3826
1.97e-95
465
644
3
184
Protein of unknown function (DUF3826). This is a putative sugar-binding family.
more
Hit ID
E-Value
Query Start
Query End
Hit Start
Hit End
Description
6GOC_A
0.0
15
649
19
654
ChainA, DUF3826 domain-containing protein [Bacteroides thetaiotaomicron]
more
3KDW_A
4.21e-12
465
646
35
216
ChainA, Putative sugar binding protein [Phocaeicola vulgatus ATCC 8482]
more
3G6I_A
2.18e-10
465
646
17
199
Crystalstructure of an outer membrane protein, part of a putative carbohydrate binding complex (bt_1022) from bacteroides thetaiotaomicron vpi-5482 at 1.93 A resolution [Bacteroides thetaiotaomicron VPI-5482]
more
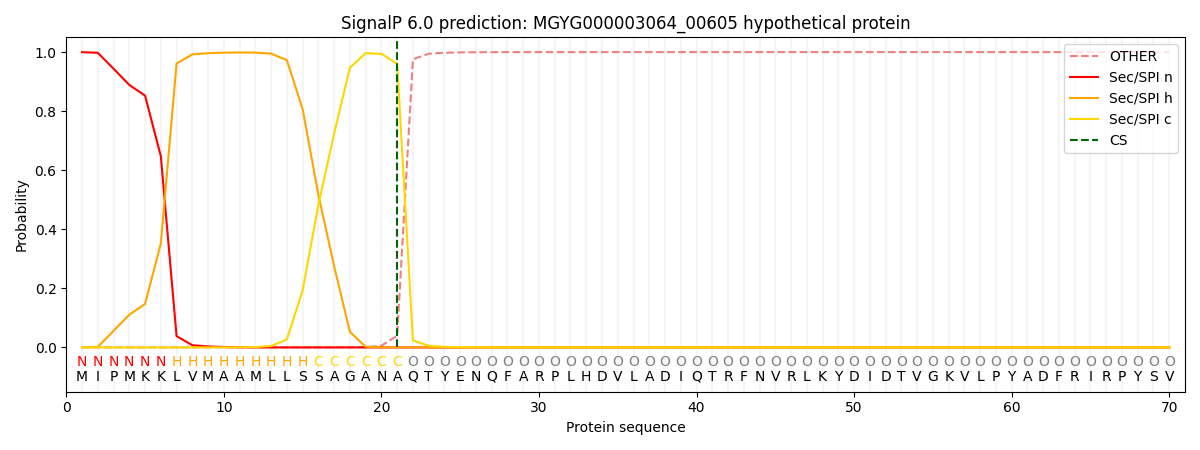
This protein is predicted as SP
Other
SP_Sec_SPI
LIPO_Sec_SPII
TAT_Tat_SPI
TATLIP_Sec_SPII
PILIN_Sec_SPIII
0.001353
0.997052
0.000649
0.000361
0.000296
0.000254
There is no transmembrane helices in MGYG000003064_00605.