You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003065_00788
You are here: Home > Sequence: MGYG000003065_00788
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Ruminococcus_B sp900544395 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Ruminococcus_B; Ruminococcus_B sp900544395 | |||||||||||
| CAZyme ID | MGYG000003065_00788 | |||||||||||
| CAZy Family | CBM32 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 25534; End: 31356 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM32 | 66 | 197 | 2.4e-22 | 0.9516129032258065 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam13306 | LRR_5 | 9.76e-30 | 1341 | 1458 | 1 | 122 | Leucine rich repeats (6 copies). This family includes a number of leucine rich repeats. This family contains a large number of BSPA-like surface antigens from Trichomonas vaginalis. |
| sd00036 | LRR_3 | 2.89e-21 | 1362 | 1467 | 3 | 113 | leucine-rich repeats. A leucine-rich repeat (LRR) is a structural protein motif of 20-30 amino acids that is unusually rich in the hydrophobic amino acid leucine. The conserved eleven-residue sequence motif (LxxLxLxxN/CxL) within the LRRs corresponds to the beta-strand and adjacent loop regions, whereas the remaining parts of the repeats are variable. LRRs fold together to form a solenoid protein domain, termed leucine-rich repeat domain. Leucine-rich repeats are usually involved in protein-protein interactions. |
| sd00036 | LRR_3 | 9.16e-21 | 1339 | 1456 | 1 | 125 | leucine-rich repeats. A leucine-rich repeat (LRR) is a structural protein motif of 20-30 amino acids that is unusually rich in the hydrophobic amino acid leucine. The conserved eleven-residue sequence motif (LxxLxLxxN/CxL) within the LRRs corresponds to the beta-strand and adjacent loop regions, whereas the remaining parts of the repeats are variable. LRRs fold together to form a solenoid protein domain, termed leucine-rich repeat domain. Leucine-rich repeats are usually involved in protein-protein interactions. |
| pfam00754 | F5_F8_type_C | 6.26e-19 | 64 | 197 | 1 | 127 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| pfam13306 | LRR_5 | 2.74e-17 | 1384 | 1467 | 1 | 86 | Leucine rich repeats (6 copies). This family includes a number of leucine rich repeats. This family contains a large number of BSPA-like surface antigens from Trichomonas vaginalis. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QHB24341.1 | 0.0 | 3 | 1937 | 2 | 1751 |
| QEI31850.1 | 0.0 | 3 | 1937 | 2 | 1751 |
| QRT30606.1 | 0.0 | 3 | 1937 | 2 | 1751 |
| QHB23133.1 | 1.58e-96 | 1 | 1937 | 32 | 1718 |
| QEI30621.1 | 1.58e-96 | 1 | 1937 | 32 | 1718 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4A41_A | 2.70e-11 | 56 | 200 | 25 | 160 | CpGH89CBM32-5,from Clostridium perfringens, in complex with galactose [Clostridium perfringens],4A44_A CpGH89CBM32-5, from Clostridium perfringens, in complex with the Tn Antigen [Clostridium perfringens],4A45_A CpGH89CBM32-5, from Clostridium perfringens, in complex with GalNAc- beta-1,3-galactose [Clostridium perfringens],4AAX_A CpGH89CBM32-5, from Clostridium perfringens, in complex with N- acetylgalactosamine [Clostridium perfringens] |
| 2ZXQ_A | 2.45e-06 | 68 | 197 | 1233 | 1369 | Crystalstructure of endo-alpha-N-acetylgalactosaminidase from Bifidobacterium longum (EngBF) [Bifidobacterium longum] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
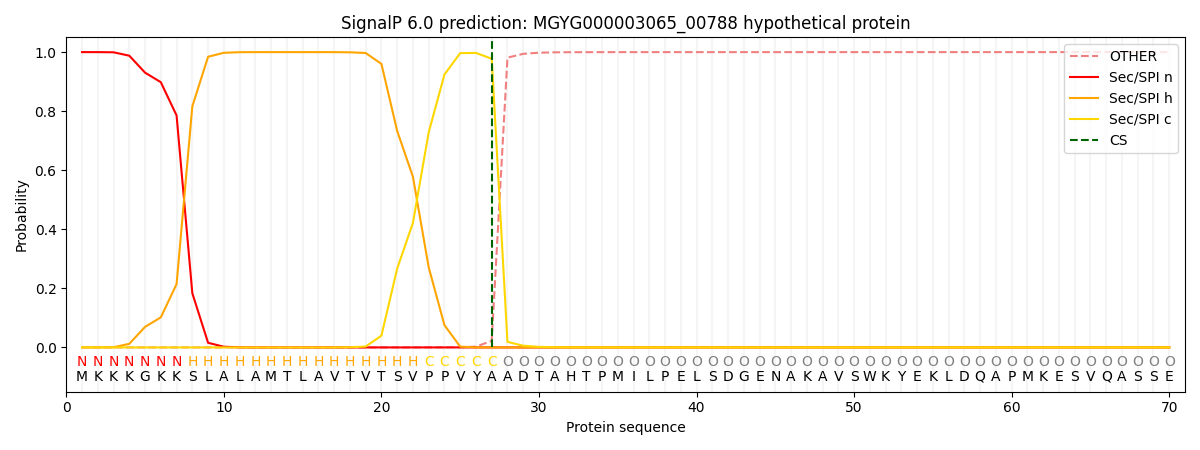
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000226 | 0.999076 | 0.000169 | 0.000175 | 0.000156 | 0.000135 |
