You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003068_00255
You are here: Home > Sequence: MGYG000003068_00255
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
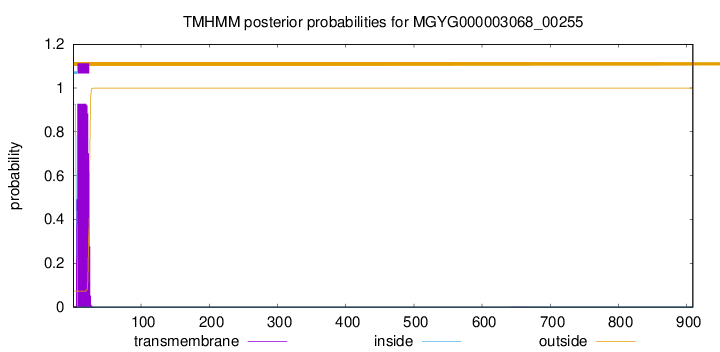
TMHMM annotations
Basic Information help
| Species | UMGS1388 sp900551345 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Monoglobales; UMGS1388; UMGS1388; UMGS1388 sp900551345 | |||||||||||
| CAZyme ID | MGYG000003068_00255 | |||||||||||
| CAZy Family | PL29 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 9540; End: 12269 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL29 | 74 | 363 | 4.2e-28 | 0.9169435215946844 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| NF033679 | DNRLRE_dom | 2.44e-24 | 758 | 908 | 1 | 164 | DNRLRE domain. The DNRLRE domain, with a length of about 160 amino acids, appears typically in large, repetitive surface proteins of bacteria and archaea, sometimes repeated several times. It occurs, notably, three times in the C-terminal region of the enzyme disaggregatase from the archaeal species Methanosarcina mazei, each time with the motif DNRLRE, for which the domain is named. Archaeal proteins within this family are described particularly well by the currently more narrowly defined Pfam model, PF06848. Note that the catalytic region of disaggregatase, in the N-terminal portion of the protein, is modeled by a different HMM, PF08480. |
| pfam13205 | Big_5 | 3.63e-09 | 546 | 644 | 4 | 105 | Bacterial Ig-like domain. |
| COG5434 | Pgu1 | 3.43e-08 | 69 | 116 | 77 | 124 | Polygalacturonase [Carbohydrate transport and metabolism]. |
| pfam12708 | Pectate_lyase_3 | 8.51e-08 | 76 | 168 | 3 | 106 | Pectate lyase superfamily protein. This family of proteins possesses a beta helical structure like Pectate lyase. This family is most closely related to glycosyl hydrolase family 28. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AQT68105.1 | 1.33e-150 | 30 | 575 | 151 | 693 |
| ATB28457.1 | 1.08e-131 | 27 | 541 | 27 | 542 |
| QRO01698.1 | 6.33e-130 | 27 | 541 | 27 | 542 |
| AFE03983.1 | 1.36e-127 | 25 | 543 | 22 | 542 |
| ADO74407.1 | 4.54e-125 | 27 | 541 | 27 | 542 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5LW3_A | 8.65e-06 | 73 | 147 | 1 | 75 | Azotobactervinelandii Mannuronan C-5 epimerase AlgE6 A-module [Azotobacter vinelandii] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q9ZFG9 | 1.06e-07 | 73 | 147 | 1 | 75 | Alginate lyase 7 OS=Azotobacter vinelandii OX=354 GN=algE7 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
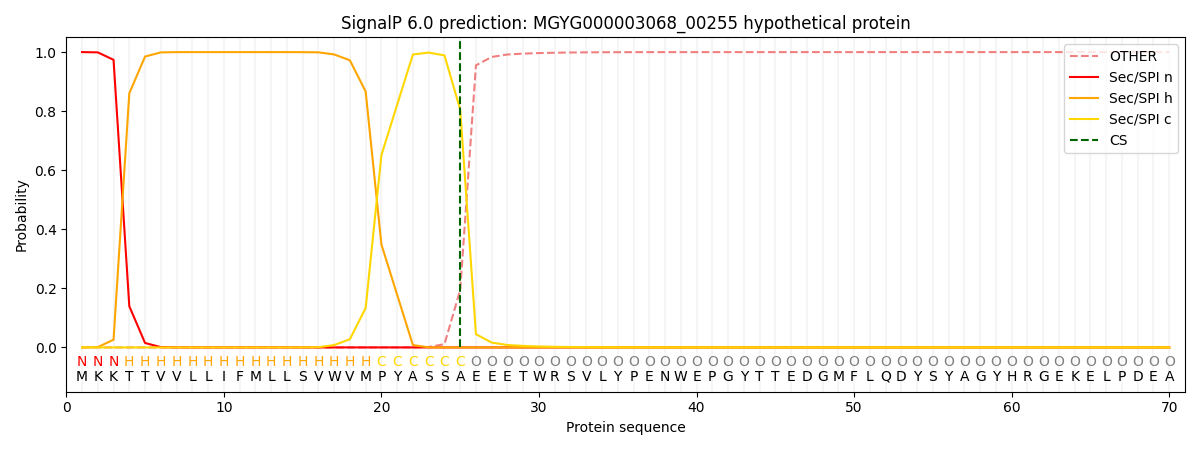
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000362 | 0.998927 | 0.000234 | 0.000160 | 0.000155 | 0.000147 |