You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003072_01045
You are here: Home > Sequence: MGYG000003072_01045
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Paenibacillus amylolyticus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Paenibacillales; Paenibacillaceae; Paenibacillus; Paenibacillus amylolyticus | |||||||||||
| CAZyme ID | MGYG000003072_01045 | |||||||||||
| CAZy Family | PL1 | |||||||||||
| CAZyme Description | Pectate lyase B | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 104446; End: 105873 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL1 | 129 | 349 | 1.8e-42 | 0.801980198019802 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| smart00656 | Amb_all | 6.79e-28 | 121 | 318 | 16 | 160 | Amb_all domain. |
| COG3866 | PelB | 1.07e-19 | 121 | 318 | 93 | 245 | Pectate lyase [Carbohydrate transport and metabolism]. |
| pfam00544 | Pec_lyase_C | 1.49e-16 | 133 | 318 | 41 | 185 | Pectate lyase. This enzyme forms a right handed beta helix structure. Pectate lyase is an enzyme involved in the maceration and soft rotting of plant tissue. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| APO42893.1 | 0.0 | 1 | 475 | 1 | 475 |
| QZN76855.1 | 0.0 | 1 | 475 | 1 | 475 |
| QOS81691.1 | 0.0 | 1 | 475 | 1 | 475 |
| QLG37876.1 | 0.0 | 1 | 475 | 1 | 475 |
| ADB78775.2 | 1.51e-313 | 1 | 475 | 1 | 475 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5GT5_A | 7.89e-294 | 31 | 475 | 2 | 446 | Structuralbasis of the specific activity and thermostability of pectate lyase (pelN) from Paenibacillus sp. 0602 [Paenibacillus sp. 0602],5GT5_B Structural basis of the specific activity and thermostability of pectate lyase (pelN) from Paenibacillus sp. 0602 [Paenibacillus sp. 0602] |
| 3ZSC_A | 1.34e-17 | 132 | 318 | 71 | 210 | Catalyticfunction and substrate recognition of the pectate lyase from Thermotoga maritima [Thermotoga maritima] |
| 1BN8_A | 8.65e-15 | 12 | 318 | 7 | 313 | BacillusSubtilis Pectate Lyase [Bacillus subtilis] |
| 1VBL_A | 1.51e-14 | 127 | 318 | 131 | 298 | Structureof the thermostable pectate lyase PL 47 [Bacillus sp. TS-47] |
| 2BSP_A | 2.05e-14 | 12 | 318 | 7 | 313 | ChainA, PROTEIN (PECTATE LYASE) [Bacillus subtilis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| D3JTC2 | 3.59e-246 | 1 | 386 | 1 | 385 | Pectate lyase B OS=Paenibacillus amylolyticus OX=1451 GN=pelB PE=1 SV=1 |
| Q9WYR4 | 2.19e-17 | 132 | 318 | 98 | 237 | Pectate trisaccharide-lyase OS=Thermotoga maritima (strain ATCC 43589 / DSM 3109 / JCM 10099 / NBRC 100826 / MSB8) OX=243274 GN=pelA PE=1 SV=1 |
| B1L969 | 3.88e-17 | 132 | 318 | 96 | 235 | Pectate trisaccharide-lyase OS=Thermotoga sp. (strain RQ2) OX=126740 GN=pelA PE=3 SV=1 |
| P39116 | 4.73e-14 | 12 | 318 | 7 | 313 | Pectate lyase OS=Bacillus subtilis (strain 168) OX=224308 GN=pel PE=1 SV=1 |
| P72242 | 1.62e-12 | 11 | 318 | 8 | 277 | Pectate lyase OS=Pseudomonas amygdali pv. lachrymans OX=53707 GN=pelP PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
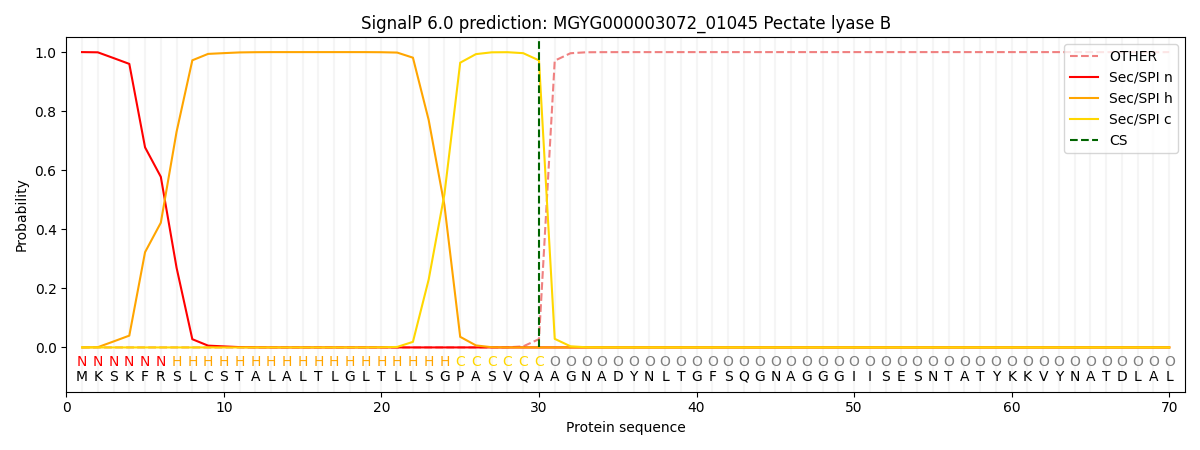
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000750 | 0.998104 | 0.000299 | 0.000307 | 0.000286 | 0.000255 |
