You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003072_01192
You are here: Home > Sequence: MGYG000003072_01192
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
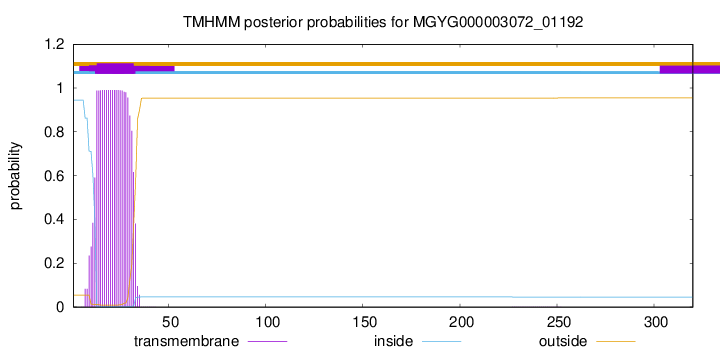
TMHMM annotations
Basic Information help
| Species | Paenibacillus amylolyticus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Paenibacillales; Paenibacillaceae; Paenibacillus; Paenibacillus amylolyticus | |||||||||||
| CAZyme ID | MGYG000003072_01192 | |||||||||||
| CAZy Family | GH10 | |||||||||||
| CAZyme Description | Endo-1,4-beta-xylanase B | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 265775; End: 266737 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH10 | 46 | 314 | 4.2e-78 | 0.9108910891089109 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3693 | XynA | 6.69e-107 | 11 | 320 | 8 | 345 | Endo-1,4-beta-xylanase, GH35 family [Carbohydrate transport and metabolism]. |
| pfam00331 | Glyco_hydro_10 | 2.64e-91 | 36 | 314 | 19 | 310 | Glycosyl hydrolase family 10. |
| smart00633 | Glyco_10 | 1.98e-84 | 65 | 312 | 2 | 263 | Glycosyl hydrolase family 10. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QZN76982.1 | 4.54e-241 | 1 | 320 | 1 | 320 |
| APO43007.1 | 8.80e-239 | 1 | 320 | 1 | 320 |
| QOS81827.1 | 7.45e-231 | 1 | 320 | 1 | 320 |
| QLG37741.1 | 5.19e-227 | 3 | 320 | 5 | 322 |
| QKS57705.1 | 2.56e-222 | 1 | 320 | 1 | 320 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4PMY_A | 3.64e-94 | 33 | 320 | 5 | 303 | Crystalstructure of GH10 endo-b-1,4-xylanase (XynB) from Xanthomonas axonopodis pv citri complexed with xylose [Xanthomonas citri pv. citri str. 306],4PMY_B Crystal structure of GH10 endo-b-1,4-xylanase (XynB) from Xanthomonas axonopodis pv citri complexed with xylose [Xanthomonas citri pv. citri str. 306],4PMZ_A Crystal structure of GH10 endo-b-1,4-xylanase (XynB) from Xanthomonas axonopodis pv citri complexed with xylobiose [Xanthomonas citri pv. citri str. 306],4PMZ_B Crystal structure of GH10 endo-b-1,4-xylanase (XynB) from Xanthomonas axonopodis pv citri complexed with xylobiose [Xanthomonas citri pv. citri str. 306] |
| 4PN2_A | 3.88e-94 | 33 | 320 | 6 | 304 | Crystalstructure of GH10 endo-b-1,4-xylanase (XynB) from Xanthomonas axonopodis pv citri complexed with xylotriose [Xanthomonas citri pv. citri str. 306],4PN2_B Crystal structure of GH10 endo-b-1,4-xylanase (XynB) from Xanthomonas axonopodis pv citri complexed with xylotriose [Xanthomonas citri pv. citri str. 306] |
| 4PMX_A | 4.01e-94 | 33 | 320 | 6 | 304 | Crystalstructure of GH10 endo-b-1,4-xylanase (XynB) from Xanthomonas axonopodis pv citri in the native form [Xanthomonas citri pv. citri str. 306] |
| 1NQ6_A | 1.03e-34 | 30 | 295 | 7 | 279 | CrystalStructure of the catalytic domain of xylanase A from Streptomyces halstedii JM8 [Streptomyces halstedii] |
| 1ISV_A | 7.81e-32 | 27 | 316 | 4 | 300 | Crystalstructure of xylanase from Streptomyces olivaceoviridis E-86 complexed with xylose [Streptomyces olivaceoviridis],1ISV_B Crystal structure of xylanase from Streptomyces olivaceoviridis E-86 complexed with xylose [Streptomyces olivaceoviridis],1ISW_A Crystal structure of xylanase from Streptomyces olivaceoviridis E-86 complexed with xylobiose [Streptomyces olivaceoviridis],1ISW_B Crystal structure of xylanase from Streptomyces olivaceoviridis E-86 complexed with xylobiose [Streptomyces olivaceoviridis],1ISX_A Crystal structure of xylanase from Streptomyces olivaceoviridis E-86 complexed with xylotriose [Streptomyces olivaceoviridis],1ISX_B Crystal structure of xylanase from Streptomyces olivaceoviridis E-86 complexed with xylotriose [Streptomyces olivaceoviridis],1ISY_A Crystal structure of xylanase from Streptomyces olivaceoviridis E-86 complexed with glucose [Streptomyces olivaceoviridis],1ISY_B Crystal structure of xylanase from Streptomyces olivaceoviridis E-86 complexed with glucose [Streptomyces olivaceoviridis],1ISZ_A Crystal structure of xylanase from Streptomyces olivaceoviridis E-86 complexed with galactose [Streptomyces olivaceoviridis],1ISZ_B Crystal structure of xylanase from Streptomyces olivaceoviridis E-86 complexed with galactose [Streptomyces olivaceoviridis],1IT0_A Crystal structure of xylanase from Streptomyces olivaceoviridis E-86 complexed with lactose [Streptomyces olivaceoviridis],1IT0_B Crystal structure of xylanase from Streptomyces olivaceoviridis E-86 complexed with lactose [Streptomyces olivaceoviridis],1V6U_A Crystal Structure Of Xylanase From Streptomyces Olivaceoviridis E-86 Complexed With 2(2)-alpha-L-arabinofuranosyl-xylobiose [Streptomyces olivaceoviridis],1V6U_B Crystal Structure Of Xylanase From Streptomyces Olivaceoviridis E-86 Complexed With 2(2)-alpha-L-arabinofuranosyl-xylobiose [Streptomyces olivaceoviridis],1V6V_A Crystal Structure Of Xylanase From Streptomyces Olivaceoviridis E-86 Complexed With 3(2)-alpha-L-arabinofuranosyl-xylotriose [Streptomyces olivaceoviridis],1V6V_B Crystal Structure Of Xylanase From Streptomyces Olivaceoviridis E-86 Complexed With 3(2)-alpha-L-arabinofuranosyl-xylotriose [Streptomyces olivaceoviridis],1V6W_A Crystal Structure Of Xylanase From Streptomyces Olivaceoviridis E-86 Complexed With 2(2)-4-O-methyl-alpha-D-glucuronosyl-xylobiose [Streptomyces olivaceoviridis],1V6W_B Crystal Structure Of Xylanase From Streptomyces Olivaceoviridis E-86 Complexed With 2(2)-4-O-methyl-alpha-D-glucuronosyl-xylobiose [Streptomyces olivaceoviridis],1V6X_A Crystal Structure Of Xylanase From Streptomyces Olivaceoviridis E-86 Complexed With 3(3)-4-O-methyl-alpha-D-glucuronosyl-xylotriose [Streptomyces olivaceoviridis],1V6X_B Crystal Structure Of Xylanase From Streptomyces Olivaceoviridis E-86 Complexed With 3(3)-4-O-methyl-alpha-D-glucuronosyl-xylotriose [Streptomyces olivaceoviridis],1XYF_A Endo-1,4-Beta-Xylanase From Streptomyces Olivaceoviridis [Streptomyces olivaceoviridis],1XYF_B Endo-1,4-Beta-Xylanase From Streptomyces Olivaceoviridis [Streptomyces olivaceoviridis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P23030 | 6.60e-70 | 37 | 316 | 323 | 597 | Endo-1,4-beta-xylanase B OS=Cellvibrio japonicus (strain Ueda107) OX=498211 GN=xynB PE=1 SV=2 |
| O94163 | 5.17e-32 | 12 | 296 | 15 | 305 | Endo-1,4-beta-xylanase F1 OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=xynF1 PE=1 SV=1 |
| Q4P902 | 1.93e-31 | 46 | 308 | 66 | 331 | Endo-1,4-beta-xylanase UM03411 OS=Ustilago maydis (strain 521 / FGSC 9021) OX=237631 GN=UMAG_03411 PE=1 SV=1 |
| C5J411 | 5.27e-31 | 33 | 316 | 37 | 327 | Probable endo-1,4-beta-xylanase C OS=Aspergillus niger OX=5061 GN=xlnC PE=2 SV=2 |
| A2QFV7 | 1.02e-30 | 33 | 316 | 37 | 327 | Probable endo-1,4-beta-xylanase C OS=Aspergillus niger (strain CBS 513.88 / FGSC A1513) OX=425011 GN=xlnC PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
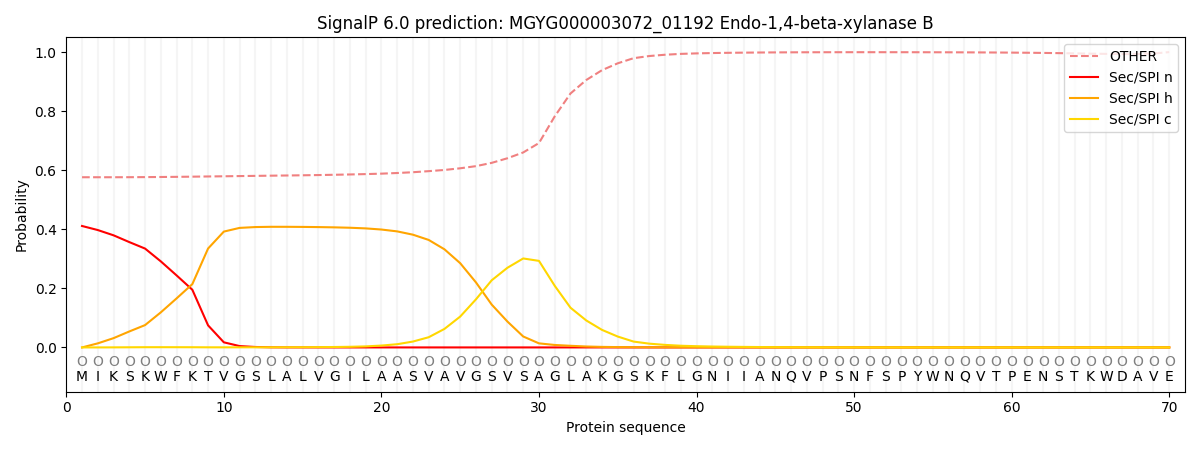
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.596599 | 0.385536 | 0.009176 | 0.001606 | 0.000782 | 0.006308 |