You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003072_03038
You are here: Home > Sequence: MGYG000003072_03038
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
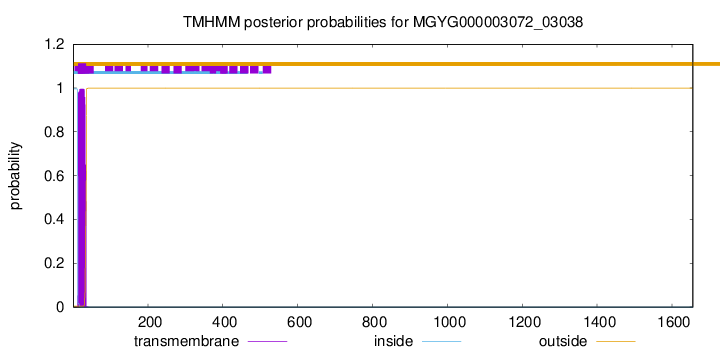
TMHMM annotations
Basic Information help
| Species | Paenibacillus amylolyticus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Paenibacillales; Paenibacillaceae; Paenibacillus; Paenibacillus amylolyticus | |||||||||||
| CAZyme ID | MGYG000003072_03038 | |||||||||||
| CAZy Family | CBM32 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 7333; End: 12303 Strand: + | |||||||||||
Full Sequence Download help
| MKVRTSQRGK TRNIYMNLLL SVLLLISSIP LWPVSKVNAA DEEHHLLSLN RPVYSSSSLG | 60 |
| GNTADLVVDG NKNSRWESIW QQDPQWIYID LGTVASISGI SIQWENAYAS AFDLEVSNDE | 120 |
| VNWQSVYSTT KGQGGLTEIE VTAEARYVRL FSHQRAQPAY GVSVYEFNVY GTGGANPPPK | 180 |
| PLAMNLALGQ PVTASSEEID EPSRSAEDKA KMEKGNYEAR NVTDGRPDTR WSSIYKDQEW | 240 |
| IVVDLGLIRE IGNVSLQWEN AFGRAYDIQV SNDADQWTTL YRELHSDGGR DEIPVYAEAR | 300 |
| YVKLAGLGRG TTNGYSLYAF DVYEYIEGDA KPVHTIPNIP EPSSVKVGSG SYAVNDITML | 360 |
| QPKNPKNRTA DITAPLPSND WWQSILVSDL GDGNSLVTLP LKSRYTKQGL QLLNPGAGYV | 420 |
| SADGGSMDAD GEPDLIMSTS NMNPAEVNTK VAGYGDYSAT IIMSDDDTAK MNTTFVKGSP | 480 |
| FLYNTFDNPD TIIVRSPNIT RLFDDQNREI VLNDGESLTA DHIGVEVTNQ DRAPTPQTFT | 540 |
| RNYGIFAPEG TVFMKLGNTL KIKLGQSEDY LSLATLPSAA ELSYYYRHAY AFVTDTTVDY | 600 |
| NYNESTSLVT TTFNSVTRTK RAGFSSETVM ALLPHQWKLA TTPVTELAYP SIRGMMKVSE | 660 |
| GNTFTTQDRF YGIIPQFVEP DDPTYSRAQL ISYLDQLDAD TAGNLMSEDP YWQGKKLHPL | 720 |
| ALGVLISDQI GDTVRRDNYL SLLRTILTDW YTYSPDEPLH SYYFYYSDEW GTIFPYGSGF | 780 |
| GVNTGLTDHH FTYGYYVFAS AVLATYDKTF LQEYGDMVEL LIRDYANPSR TDDQFPWLRN | 840 |
| FDPYAGHSWA GGYADNRSGN NQEAAGEALF SWVGQYMWGE VTGNKAYRDT GIWGFMTEEK | 900 |
| AAEQYWFNYD GDNWLEDYQH ATVGHVYGSA YLYGTYFSNE AEHIYGIHWL PPAEWMTYYG | 960 |
| RDPQNTKALY NGLIKDLGGQ QERTWQHIIW PFQSIGDPQG ALAKWNTSEM QQNEVFNAYW | 1020 |
| FMHSMASTGT RTMDIWADDP AATVYEKDGV YTAQIWNPSD TVKTVHFYNA HGALGSATVY | 1080 |
| AKAQVKVNPL EHSVVKQPDA SQGVVYLDRS QWTITASSSS EPVERMVDGD LTTRWSSGQI | 1140 |
| QKPGDWLHID LGGEQFMDTV FMNSGNNGGD YAHGYEIYVS KDNENWSEPI AEGAGTSPSL | 1200 |
| SVDLGMQTAR YVKIVLTSSA DSWWSISELK LARFGKLEAT PELPQPVPPP DRSSWIVTAS | 1260 |
| STQGADVTAN MLDGSSDTLW TNGRKQTKGQ WITIDLGQKS FFDAIELDPG HAKVDYPRQY | 1320 |
| RIYVSDDGEH WGTSIAGGEG TSGRMSITFP EQQARYVKMV QIGDSDQWWT ISELFVHHNG | 1380 |
| TGKQKRLLPN GWSVTASSGD DAAAILDGSD QTRWTSGQTQ TGGEWIQLDL GASQPVNRIV | 1440 |
| LDSAHSREDF ARGYEIYTSL DGEHWGKALA SGEGSDALLS IAFTPHEARY IKVVQTGTDS | 1500 |
| HWWSVAELEV YTADQTPWIS GEQDHLLPAE LDRTSWTATA STSEDVAAML DDDLNSRWTS | 1560 |
| SSGQYANQWV QIDLGSKQSF SRLTMDSGSS SNDYARNFQF FTSEDGERWE GVTEALGTGP | 1620 |
| VISHTFSRQT ARYIKIALTA DTPEWWWSIA ELKLYH | 1656 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH81 | 362 | 1020 | 1.2e-126 | 0.977491961414791 |
| CBM32 | 55 | 168 | 2.2e-24 | 0.9193548387096774 |
| CBM32 | 1116 | 1230 | 2.8e-22 | 0.9032258064516129 |
| CBM32 | 1259 | 1374 | 3.3e-22 | 0.9032258064516129 |
| CBM32 | 1396 | 1509 | 4.3e-22 | 0.8870967741935484 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG5498 | Acf2 | 1.18e-63 | 342 | 1008 | 42 | 732 | Endoglucanase Acf2 [Carbohydrate transport and metabolism]. |
| pfam17652 | Glyco_hydro81C | 5.23e-38 | 764 | 1013 | 81 | 337 | Glycosyl hydrolase family 81 C-terminal domain. Family of eukaryotic beta-1,3-glucanases. Within the Aspergillus fumigatus protein, two perfectly conserved Glu residues (E550 or E554) have been proposed as putative nucleophiles of the active site of the Engl1 endoglucanase, while the proton donor would be D475. The endo-beta-1,3-glucanase activity is essential for efficient spore release. This entry represents the helical C-terminal domain. |
| pfam00754 | F5_F8_type_C | 2.86e-17 | 55 | 167 | 5 | 127 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| pfam00754 | F5_F8_type_C | 1.06e-16 | 215 | 319 | 10 | 125 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| pfam00754 | F5_F8_type_C | 6.12e-16 | 1257 | 1374 | 2 | 127 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QZN75609.1 | 0.0 | 1 | 1656 | 1 | 1656 |
| ANS76332.1 | 0.0 | 8 | 1656 | 8 | 1663 |
| ASS68372.1 | 0.0 | 39 | 1377 | 38 | 1403 |
| QJC52720.1 | 0.0 | 2 | 1377 | 3 | 1391 |
| CDG84341.1 | 0.0 | 45 | 1235 | 51 | 1239 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6FOP_A | 2.97e-107 | 348 | 1067 | 6 | 698 | ChainA, Glycoside hydrolase family 81 [Acetivibrio thermocellus ATCC 27405] |
| 5V1W_A | 1.22e-82 | 344 | 1071 | 3 | 738 | ChainA, Glycoside Hydrolase [Halalkalibacterium halodurans C-125] |
| 5T4A_A | 1.71e-82 | 336 | 1071 | 15 | 758 | ChainA, Glycoside Hydrolase [Halalkalibacterium halodurans C-125],5T4G_A Chain A, Glycoside Hydrolase [Halalkalibacterium halodurans C-125] |
| 5UPI_A | 3.08e-82 | 344 | 1071 | 3 | 738 | ChainA, BH0236 protein [Halalkalibacterium halodurans C-125],5UPM_A Chain A, BH0236 protein [Halalkalibacterium halodurans C-125],5UPN_A Chain A, BH0236 protein [Halalkalibacterium halodurans C-125],5UPO_A Chain A, BH0236 protein [Halalkalibacterium halodurans C-125] |
| 5T4C_A | 4.21e-82 | 336 | 1071 | 15 | 758 | ChainA, Glycoside Hydrolase [Halalkalibacterium halodurans C-125] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q9UT45 | 2.37e-28 | 625 | 1009 | 317 | 715 | Primary septum endo-1,3(4)-beta-glucanase OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=eng1 PE=3 SV=1 |
| P53753 | 9.43e-21 | 696 | 1008 | 768 | 1088 | Endo-1,3(4)-beta-glucanase 1 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=DSE4 PE=1 SV=1 |
| Q12168 | 1.78e-19 | 622 | 1029 | 346 | 776 | Endo-1,3(4)-beta-glucanase 2 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=ACF2 PE=1 SV=1 |
| D4AZ24 | 3.61e-19 | 620 | 1001 | 469 | 872 | Probable endo-1,3(4)-beta-glucanase ARB_01444 OS=Arthroderma benhamiae (strain ATCC MYA-4681 / CBS 112371) OX=663331 GN=ARB_01444 PE=1 SV=1 |
| Q09850 | 2.43e-18 | 783 | 1029 | 452 | 703 | Ascus wall endo-1,3(4)-beta-glucanase OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=eng2 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
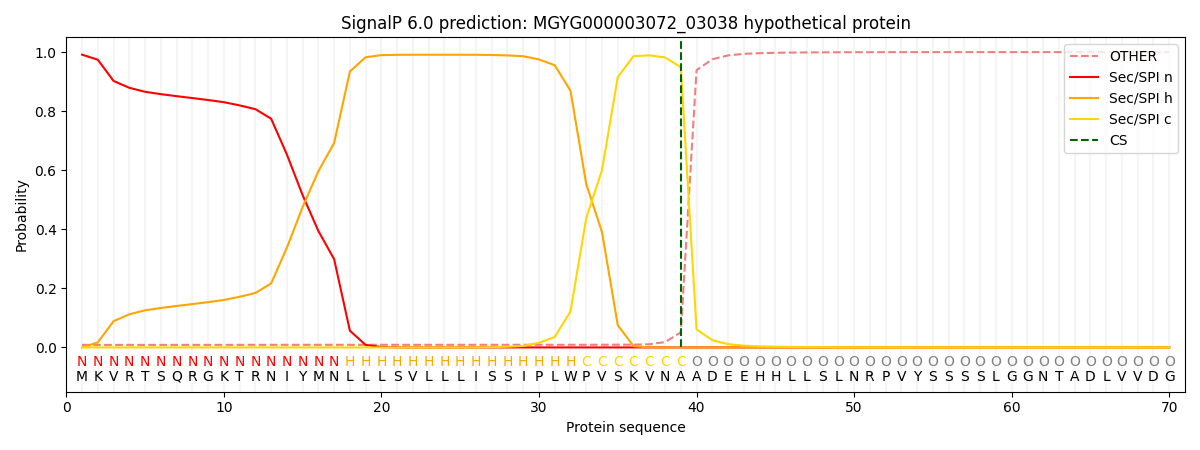
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.009968 | 0.989039 | 0.000278 | 0.000304 | 0.000198 | 0.000184 |