You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003072_03133
You are here: Home > Sequence: MGYG000003072_03133
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Paenibacillus amylolyticus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Paenibacillales; Paenibacillaceae; Paenibacillus; Paenibacillus amylolyticus | |||||||||||
| CAZyme ID | MGYG000003072_03133 | |||||||||||
| CAZy Family | GH30 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 118878; End: 120722 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH30 | 37 | 459 | 9e-152 | 0.993421052631579 |
| CBM13 | 473 | 562 | 2.4e-21 | 0.4574468085106383 |
| CBM13 | 522 | 613 | 2.9e-21 | 0.4787234042553192 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam14587 | Glyco_hydr_30_2 | 3.59e-37 | 38 | 334 | 7 | 357 | O-Glycosyl hydrolase family 30. |
| COG5520 | XynC | 4.74e-25 | 4 | 460 | 5 | 427 | O-Glycosyl hydrolase [Cell wall/membrane/envelope biogenesis]. |
| pfam14200 | RicinB_lectin_2 | 4.22e-24 | 511 | 598 | 1 | 89 | Ricin-type beta-trefoil lectin domain-like. |
| pfam14200 | RicinB_lectin_2 | 7.00e-23 | 472 | 552 | 9 | 89 | Ricin-type beta-trefoil lectin domain-like. |
| pfam00652 | Ricin_B_lectin | 4.16e-19 | 476 | 608 | 1 | 126 | Ricin-type beta-trefoil lectin domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| APO47158.1 | 0.0 | 1 | 572 | 1 | 572 |
| QZN75682.1 | 0.0 | 1 | 572 | 1 | 572 |
| QOS80535.1 | 0.0 | 1 | 572 | 1 | 572 |
| QLG39030.1 | 0.0 | 1 | 572 | 1 | 572 |
| QKS56604.1 | 0.0 | 1 | 524 | 1 | 524 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4FMV_A | 6.33e-20 | 44 | 460 | 14 | 385 | CrystalStructure Analysis of a GH30 Endoxylanase from Clostridium papyrosolvens C71 [Ruminiclostridium papyrosolvens DSM 2782] |
| 3CLW_A | 7.93e-20 | 36 | 396 | 9 | 429 | Crystalstructure of conserved exported protein from Bacteroides fragilis [Bacteroides fragilis NCTC 9343],3CLW_B Crystal structure of conserved exported protein from Bacteroides fragilis [Bacteroides fragilis NCTC 9343],3CLW_C Crystal structure of conserved exported protein from Bacteroides fragilis [Bacteroides fragilis NCTC 9343],3CLW_D Crystal structure of conserved exported protein from Bacteroides fragilis [Bacteroides fragilis NCTC 9343],3CLW_E Crystal structure of conserved exported protein from Bacteroides fragilis [Bacteroides fragilis NCTC 9343],3CLW_F Crystal structure of conserved exported protein from Bacteroides fragilis [Bacteroides fragilis NCTC 9343] |
| 6KRL_A | 2.91e-19 | 22 | 460 | 79 | 541 | Crystalstructure of GH30 xylanase B from Talaromyces cellulolyticus expressed by Pichia pastoris [Saccharomyces uvarum],6KRN_A Crystal structure of GH30 xylanase B from Talaromyces cellulolyticus expressed by Pichia pastoris in complex with aldotriuronic acid [Saccharomyces uvarum] |
| 6IUJ_A | 3.70e-19 | 21 | 460 | 6 | 470 | ChainA, GH30 Xylanase B [Talaromyces cellulolyticus CF-2612],6IUJ_B Chain B, GH30 Xylanase B [Talaromyces cellulolyticus CF-2612] |
| 6M5Z_A | 2.46e-18 | 34 | 462 | 24 | 454 | ChainA, GH30 Xylanase C [Talaromyces cellulolyticus CF-2612],6M5Z_B Chain B, GH30 Xylanase C [Talaromyces cellulolyticus CF-2612] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q76FP5 | 2.08e-84 | 34 | 463 | 23 | 475 | Endo-beta-1,6-galactanase OS=Hypocrea rufa OX=5547 GN=6GAL PE=1 SV=1 |
| A0A401ETL2 | 7.41e-24 | 21 | 503 | 18 | 676 | Exo-beta-1,6-galactobiohydrolase OS=Bifidobacterium longum subsp. longum (strain ATCC 15707 / DSM 20219 / JCM 1217 / NCTC 11818 / E194b) OX=565042 GN=bl1,6Gal PE=1 SV=1 |
| Q45070 | 1.58e-08 | 10 | 462 | 12 | 421 | Glucuronoxylanase XynC OS=Bacillus subtilis (strain 168) OX=224308 GN=xynC PE=1 SV=1 |
| Q6YK37 | 3.49e-07 | 166 | 461 | 138 | 421 | Glucuronoxylanase XynC OS=Bacillus subtilis OX=1423 GN=xynC PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
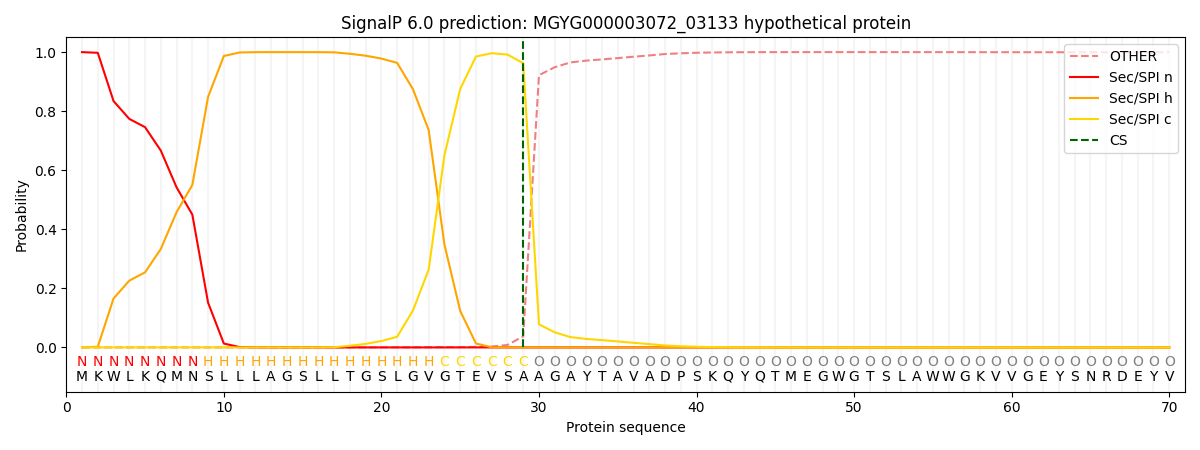
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000781 | 0.997615 | 0.000614 | 0.000415 | 0.000289 | 0.000264 |
