You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003076_01722
You are here: Home > Sequence: MGYG000003076_01722
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Marinifilaceae; Odoribacter; | |||||||||||
| CAZyme ID | MGYG000003076_01722 | |||||||||||
| CAZy Family | GH50 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 2670; End: 3272 Strand: + | |||||||||||
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QNL39155.1 | 1.87e-134 | 1 | 200 | 1 | 200 |
| QGT71933.1 | 2.66e-134 | 1 | 200 | 1 | 200 |
| QRN00280.1 | 1.86e-129 | 1 | 200 | 1 | 200 |
| QUT30898.1 | 1.86e-129 | 1 | 200 | 1 | 200 |
| CBK66219.1 | 1.86e-129 | 1 | 200 | 1 | 200 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5T3B_A | 4.79e-26 | 51 | 200 | 24 | 167 | ChainA, Glycoside Hydrolase [Phocaeicola plebeius],5T3B_B Chain B, Glycoside Hydrolase [Phocaeicola plebeius] |
| 4BQ2_A | 4.33e-06 | 113 | 192 | 243 | 315 | Structuralanalysis of an exo-beta-agarase [Saccharophagus degradans 2-40],4BQ2_B Structural analysis of an exo-beta-agarase [Saccharophagus degradans 2-40],4BQ2_C Structural analysis of an exo-beta-agarase [Saccharophagus degradans 2-40],4BQ2_D Structural analysis of an exo-beta-agarase [Saccharophagus degradans 2-40],4BQ3_A Structural analysis of an exo-beta-agarase [Saccharophagus degradans 2-40],4BQ3_B Structural analysis of an exo-beta-agarase [Saccharophagus degradans 2-40],4BQ3_C Structural analysis of an exo-beta-agarase [Saccharophagus degradans 2-40],4BQ3_D Structural analysis of an exo-beta-agarase [Saccharophagus degradans 2-40] |
| 4BQ4_A | 4.33e-06 | 113 | 192 | 243 | 315 | Structuralanalysis of an exo-beta-agarase [Saccharophagus degradans 2-40],4BQ4_B Structural analysis of an exo-beta-agarase [Saccharophagus degradans 2-40],4BQ5_A Structural analysis of an exo-beta-agarase [Saccharophagus degradans 2-40],4BQ5_B Structural analysis of an exo-beta-agarase [Saccharophagus degradans 2-40] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as LIPO
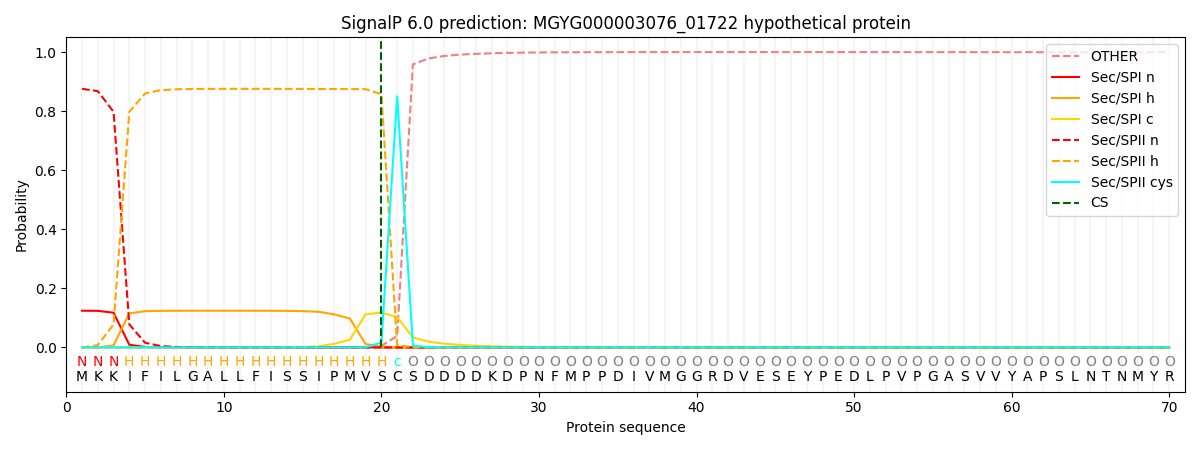
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000269 | 0.120793 | 0.878774 | 0.000060 | 0.000074 | 0.000059 |
