You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003087_01773
You are here: Home > Sequence: MGYG000003087_01773
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
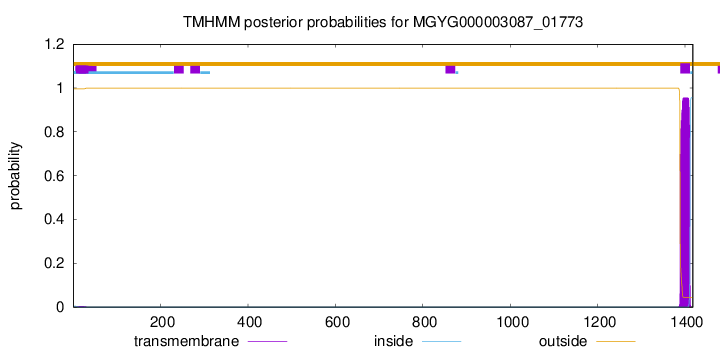
TMHMM annotations
Basic Information help
| Species | Merdibacter sp900543035 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Erysipelotrichales; Erysipelotrichaceae; Merdibacter; Merdibacter sp900543035 | |||||||||||
| CAZyme ID | MGYG000003087_01773 | |||||||||||
| CAZy Family | GH84 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 19962; End: 24218 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH84 | 191 | 483 | 6.5e-103 | 0.9932203389830508 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam07555 | NAGidase | 1.54e-130 | 191 | 482 | 1 | 293 | beta-N-acetylglucosaminidase. This family has previously been described as a hyaluronidase. However, more recently it has been shown that this family has beta-N-acetylglucosaminidase activity. |
| pfam02838 | Glyco_hydro_20b | 2.18e-10 | 42 | 184 | 3 | 123 | Glycosyl hydrolase family 20, domain 2. This domain has a zincin-like fold. |
| pfam00754 | F5_F8_type_C | 1.31e-09 | 770 | 898 | 9 | 127 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| TIGR02168 | SMC_prok_B | 4.69e-07 | 1019 | 1319 | 762 | 1041 | chromosome segregation protein SMC, common bacterial type. SMC (structural maintenance of chromosomes) proteins bind DNA and act in organizing and segregating chromosomes for partition. SMC proteins are found in bacteria, archaea, and eukaryotes. This family represents the SMC protein of most bacteria. The smc gene is often associated with scpB (TIGR00281) and scpA genes, where scp stands for segregation and condensation protein. SMC was shown (in Caulobacter crescentus) to be induced early in S phase but present and bound to DNA throughout the cell cycle. [Cellular processes, Cell division, DNA metabolism, Chromosome-associated proteins] |
| COG5281 | COG5281 | 8.43e-04 | 963 | 1370 | 78 | 475 | Phage-related minor tail protein [Mobilome: prophages, transposons]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AYE33733.1 | 0.0 | 1 | 1237 | 1 | 1239 |
| QAS61890.1 | 0.0 | 1 | 1237 | 1 | 1239 |
| ATD57955.1 | 0.0 | 1 | 1231 | 1 | 1240 |
| SLK13973.1 | 0.0 | 1 | 1231 | 1 | 1240 |
| QBJ74644.1 | 0.0 | 1 | 1231 | 1 | 1240 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5OXD_A | 1.09e-92 | 45 | 574 | 20 | 546 | Complexof a C. perfringens O-GlcNAcase with a fragment hit [Clostridium perfringens] |
| 2J62_A | 1.21e-92 | 45 | 574 | 18 | 544 | Structureof a bacterial O-glcnacase in complex with glcnacstatin [Clostridium perfringens],2J62_B Structure of a bacterial O-glcnacase in complex with glcnacstatin [Clostridium perfringens],2WB5_A GlcNAcstatins are nanomolar inhibitors of human O-GlcNAcase inducing cellular hyper-O-GlcNAcylation [Clostridium perfringens],2WB5_B GlcNAcstatins are nanomolar inhibitors of human O-GlcNAcase inducing cellular hyper-O-GlcNAcylation [Clostridium perfringens] |
| 6PV5_A | 1.80e-92 | 31 | 569 | 32 | 572 | Structureof CpGH84B [Clostridium perfringens ATCC 13124] |
| 4ZXL_A | 3.91e-92 | 45 | 574 | 10 | 536 | CpOGAD298N in complex with Drosophila HCF -derived Thr-O-GlcNAc peptide [Clostridium perfringens ATCC 13124] |
| 2YDQ_A | 5.24e-92 | 45 | 574 | 20 | 546 | CpOGAD298N in complex with hOGA-derived O-GlcNAc peptide [Clostridium perfringens],2YDR_A CpOGA D298N in complex with p53-derived O-GlcNAc peptide [Clostridium perfringens],2YDS_A CpOGA D298N in complex with TAB1-derived O-GlcNAc peptide [Clostridium perfringens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q8XL08 | 3.72e-88 | 27 | 819 | 30 | 819 | O-GlcNAcase NagJ OS=Clostridium perfringens (strain 13 / Type A) OX=195102 GN=nagJ PE=1 SV=1 |
| Q0TR53 | 9.27e-88 | 27 | 819 | 30 | 819 | O-GlcNAcase NagJ OS=Clostridium perfringens (strain ATCC 13124 / DSM 756 / JCM 1290 / NCIMB 6125 / NCTC 8237 / Type A) OX=195103 GN=nagJ PE=1 SV=1 |
| Q89ZI2 | 2.18e-85 | 126 | 721 | 95 | 693 | O-GlcNAcase BT_4395 OS=Bacteroides thetaiotaomicron (strain ATCC 29148 / DSM 2079 / JCM 5827 / CCUG 10774 / NCTC 10582 / VPI-5482 / E50) OX=226186 GN=BT_4395 PE=1 SV=1 |
| P26831 | 3.01e-69 | 1 | 986 | 1 | 1027 | Hyaluronoglucosaminidase OS=Clostridium perfringens (strain 13 / Type A) OX=195102 GN=nagH PE=1 SV=2 |
| O60502 | 3.08e-29 | 190 | 451 | 61 | 331 | Protein O-GlcNAcase OS=Homo sapiens OX=9606 GN=OGA PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
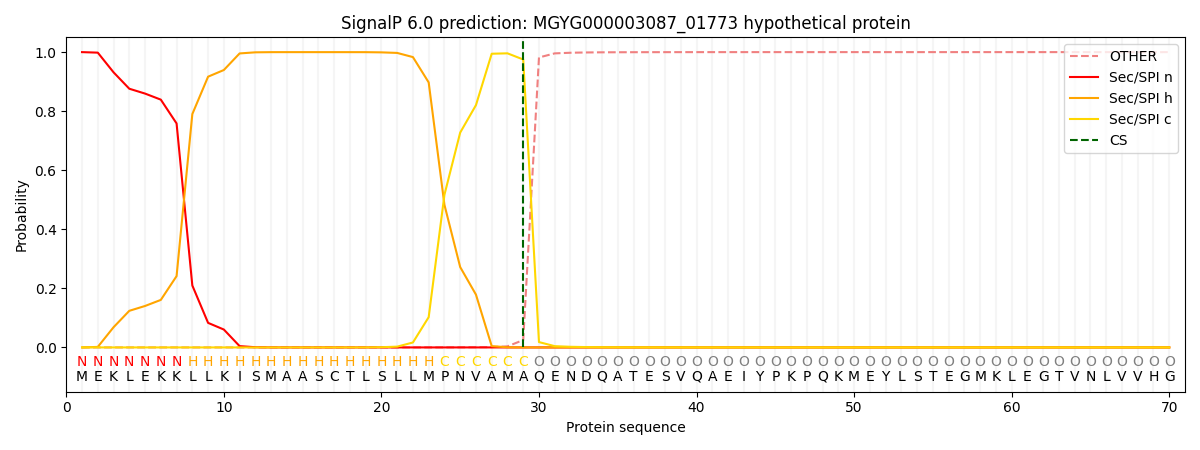
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000353 | 0.998557 | 0.000462 | 0.000228 | 0.000196 | 0.000166 |