You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003087_02017
You are here: Home > Sequence: MGYG000003087_02017
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
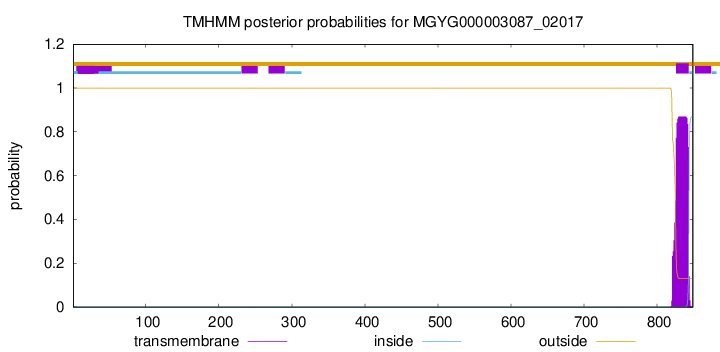
TMHMM annotations
Basic Information help
| Species | Merdibacter sp900543035 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Erysipelotrichales; Erysipelotrichaceae; Merdibacter; Merdibacter sp900543035 | |||||||||||
| CAZyme ID | MGYG000003087_02017 | |||||||||||
| CAZy Family | CBM32 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 5940; End: 8489 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd06596 | GH31_CPE1046 | 1.28e-72 | 1 | 141 | 197 | 334 | Clostridium CPE1046-like. CPE1046 is an uncharacterized Clostridium perfringens protein with a glycosyl hydrolase family 31 (GH31) domain. The domain architecture of CPE1046 and its orthologs includes a C-terminal fibronectin type 3 (FN3) domain and a coagulation factor 5/8 type C domain in addition to the GH31 domain. Enzymes of the GH31 family possess a wide range of different hydrolytic activities including alpha-glucosidase (glucoamylase and sucrase-isomaltase), alpha-xylosidase, 6-alpha-glucosyltransferase, 3-alpha-isomaltosyltransferase and alpha-1,4-glucan lyase. All GH31 enzymes cleave a terminal carbohydrate moiety from a substrate that varies considerably in size, depending on the enzyme, and may be either a starch or a glycoprotein. |
| COG1501 | YicI | 6.64e-46 | 11 | 254 | 522 | 753 | Alpha-glucosidase, glycosyl hydrolase family GH31 [Carbohydrate transport and metabolism]. |
| pfam01055 | Glyco_hydro_31 | 6.99e-45 | 13 | 170 | 295 | 442 | Glycosyl hydrolases family 31. Glycosyl hydrolases are key enzymes of carbohydrate metabolism. Family 31 comprises of enzymes that are, or similar to, alpha- galactosidases. |
| cd06603 | GH31_GANC_GANAB_alpha | 3.15e-28 | 38 | 208 | 303 | 467 | neutral alpha-glucosidase C, neutral alpha-glucosidase AB. This subgroup includes the closely related glycosyl hydrolase family 31 (GH31) isozymes, neutral alpha-glucosidase C (GANC) and the alpha subunit of heterodimeric neutral alpha-glucosidase AB (GANAB). Initially distinguished on the basis of differences in electrophoretic mobility in starch gel, GANC and GANAB have been shown to have other differences, including those of substrate specificity. GANC and GANAB are key enzymes in glycogen metabolism that hydrolyze terminal, non-reducing 1,4-linked alpha-D-glucose residues from glycogen in the endoplasmic reticulum. The GANC/GANAB family includes the alpha-glucosidase II (ModA) from Dictyostelium discoideum as well as the alpha-glucosidase II (GLS2, or ROT2 - Reversal of TOR2 lethality protein 2) from Saccharomyces cerevisiae. |
| PRK10426 | PRK10426 | 7.05e-21 | 50 | 169 | 518 | 622 | alpha-glucosidase; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QOK28184.1 | 5.66e-177 | 1 | 762 | 477 | 1262 |
| AND39696.1 | 7.92e-177 | 1 | 762 | 477 | 1262 |
| AZU64123.1 | 1.59e-171 | 1 | 761 | 486 | 1265 |
| QSW19099.1 | 3.52e-164 | 1 | 762 | 493 | 1272 |
| ATD48763.1 | 1.20e-159 | 1 | 776 | 511 | 1291 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6M76_A | 2.76e-94 | 1 | 432 | 518 | 963 | GH31alpha-N-acetylgalactosaminidase from Enterococcus faecalis [Enterococcus faecalis ATCC 10100],6M77_A GH31 alpha-N-acetylgalactosaminidase from Enterococcus faecalis in complex with N-acetylgalactosamine [Enterococcus faecalis ATCC 10100] |
| 7F7Q_A | 2.76e-94 | 1 | 432 | 518 | 963 | ChainA, GH31 alpha-N-acetylgalactosaminidase [Enterococcus faecalis ATCC 10100] |
| 7F7R_A | 2.76e-94 | 1 | 432 | 518 | 963 | ChainA, GH31 alpha-N-acetylgalactosaminidase [Enterococcus faecalis ATCC 10100] |
| 6JR8_A | 9.51e-31 | 5 | 263 | 533 | 784 | Flavobacteriumjohnsoniae GH31 dextranase, FjDex31A, mutant D412A complexed with isomaltotriose [Flavobacterium johnsoniae UW101],6JR8_B Flavobacterium johnsoniae GH31 dextranase, FjDex31A, mutant D412A complexed with isomaltotriose [Flavobacterium johnsoniae UW101],6JR8_C Flavobacterium johnsoniae GH31 dextranase, FjDex31A, mutant D412A complexed with isomaltotriose [Flavobacterium johnsoniae UW101],6JR8_D Flavobacterium johnsoniae GH31 dextranase, FjDex31A, mutant D412A complexed with isomaltotriose [Flavobacterium johnsoniae UW101] |
| 6JR6_A | 9.51e-31 | 5 | 263 | 533 | 784 | Flavobacteriumjohnsoniae GH31 dextranase, FjDex31A [Flavobacterium johnsoniae UW101],6JR6_B Flavobacterium johnsoniae GH31 dextranase, FjDex31A [Flavobacterium johnsoniae UW101],6JR6_C Flavobacterium johnsoniae GH31 dextranase, FjDex31A [Flavobacterium johnsoniae UW101],6JR6_D Flavobacterium johnsoniae GH31 dextranase, FjDex31A [Flavobacterium johnsoniae UW101],6JR7_A Flavobacterium johnsoniae GH31 dextranase, FjDex31A, complexed with glucose [Flavobacterium johnsoniae UW101],6JR7_B Flavobacterium johnsoniae GH31 dextranase, FjDex31A, complexed with glucose [Flavobacterium johnsoniae UW101],6JR7_C Flavobacterium johnsoniae GH31 dextranase, FjDex31A, complexed with glucose [Flavobacterium johnsoniae UW101],6JR7_D Flavobacterium johnsoniae GH31 dextranase, FjDex31A, complexed with glucose [Flavobacterium johnsoniae UW101] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q9F234 | 6.84e-24 | 13 | 215 | 527 | 720 | Alpha-glucosidase 2 OS=Bacillus thermoamyloliquefaciens OX=1425 PE=3 SV=1 |
| Q9P999 | 1.71e-22 | 9 | 245 | 472 | 695 | Alpha-xylosidase OS=Saccharolobus solfataricus (strain ATCC 35092 / DSM 1617 / JCM 11322 / P2) OX=273057 GN=xylS PE=1 SV=1 |
| A7LXT0 | 2.53e-20 | 11 | 270 | 692 | 941 | Alpha-xylosidase BoGH31A OS=Bacteroides ovatus (strain ATCC 8483 / DSM 1896 / JCM 5824 / BCRC 10623 / CCUG 4943 / NCTC 11153) OX=411476 GN=BACOVA_02646 PE=1 SV=1 |
| Q9FN05 | 3.84e-19 | 14 | 171 | 632 | 780 | Probable glucan 1,3-alpha-glucosidase OS=Arabidopsis thaliana OX=3702 GN=PSL5 PE=1 SV=1 |
| B9F676 | 1.15e-18 | 14 | 214 | 630 | 824 | Probable glucan 1,3-alpha-glucosidase OS=Oryza sativa subsp. japonica OX=39947 GN=Os03g0216600 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000049 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |