You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003091_01205
You are here: Home > Sequence: MGYG000003091_01205
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Parabacteroides sp900547435 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Tannerellaceae; Parabacteroides; Parabacteroides sp900547435 | |||||||||||
| CAZyme ID | MGYG000003091_01205 | |||||||||||
| CAZy Family | GH76 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 7281; End: 8378 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH76 | 37 | 352 | 3.9e-85 | 0.8547486033519553 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam03663 | Glyco_hydro_76 | 4.41e-44 | 52 | 340 | 35 | 331 | Glycosyl hydrolase family 76. Family of alpha-1,6-mannanases. |
| COG4833 | COG4833 | 4.69e-27 | 99 | 339 | 77 | 323 | Predicted alpha-1,6-mannanase, GH76 family [Carbohydrate transport and metabolism]. |
| pfam07470 | Glyco_hydro_88 | 8.20e-07 | 179 | 341 | 39 | 176 | Glycosyl Hydrolase Family 88. Unsaturated glucuronyl hydrolase catalyzes the hydrolytic release of unsaturated glucuronic acids from oligosaccharides (EC:3.2.1.-) produced by the reactions of polysaccharide lyases. |
| pfam00759 | Glyco_hydro_9 | 0.002 | 84 | 200 | 180 | 290 | Glycosyl hydrolase family 9. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QJE28248.1 | 1.06e-183 | 1 | 361 | 1 | 358 |
| QIX66371.1 | 1.14e-183 | 1 | 361 | 1 | 358 |
| AST52979.1 | 2.14e-183 | 1 | 361 | 1 | 358 |
| QUT96026.1 | 2.14e-183 | 1 | 361 | 1 | 358 |
| BBK90205.1 | 2.14e-183 | 1 | 361 | 1 | 358 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3K7X_A | 5.27e-36 | 106 | 326 | 87 | 301 | ChainA, Lin0763 protein [Listeria innocua] |
| 6SHD_A | 4.54e-24 | 38 | 326 | 75 | 351 | Structureof the GH76A alpha-1,6-mannanase from Salegentibacter sp. HEL1_6 [Salegentibacter sp. Hel_I_6],6SHD_B Structure of the GH76A alpha-1,6-mannanase from Salegentibacter sp. HEL1_6 [Salegentibacter sp. Hel_I_6],6SHD_C Structure of the GH76A alpha-1,6-mannanase from Salegentibacter sp. HEL1_6 [Salegentibacter sp. Hel_I_6] |
| 6Y8F_A | 1.04e-22 | 38 | 326 | 76 | 352 | ChainA, Alpha-1,6-endo-mannanase GH76A mutant [Salegentibacter sp. Hel_I_6] |
| 6SHM_A | 6.73e-22 | 38 | 326 | 76 | 352 | Aninactive (D136A and D137A) variant of alpha-1,6-mannanase, GH76A of Salegentibacter sp. HEL1_6 in complex with alpha-1,6-mannotetrose [Salegentibacter sp. Hel_I_6] |
| 6U4Z_A | 2.45e-18 | 45 | 356 | 161 | 468 | CrystalStructure of a family 76 glycoside hydrolase from a bovine Bacteroides thetaiotaomicron strain [Bacteroides thetaiotaomicron] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O74556 | 1.58e-19 | 47 | 339 | 64 | 359 | Putative mannan endo-1,6-alpha-mannosidase C970.02 OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=SPCC970.02 PE=3 SV=1 |
| Q9P6I3 | 2.90e-19 | 32 | 302 | 29 | 312 | Putative mannan endo-1,6-alpha-mannosidase C1198.07c OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=SPBC1198.07c PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
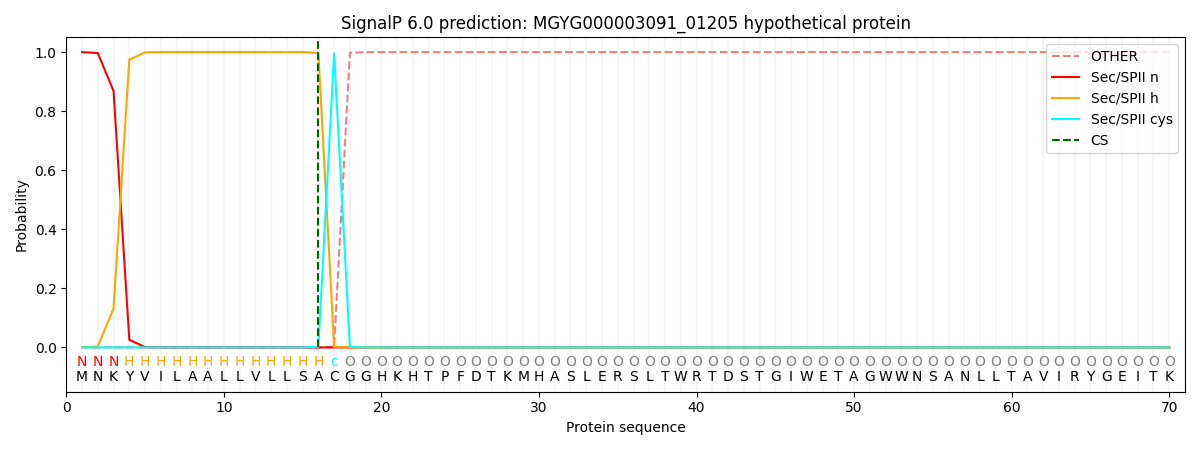
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000000 | 1.000088 | 0.000000 | 0.000000 | 0.000000 |
