You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003094_01449
You are here: Home > Sequence: MGYG000003094_01449
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Phocaeicola barnesiae | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Phocaeicola; Phocaeicola barnesiae | |||||||||||
| CAZyme ID | MGYG000003094_01449 | |||||||||||
| CAZy Family | CBM32 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 23355; End: 25154 Strand: + | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd08759 | Type_III_cohesin_like | 2.31e-21 | 476 | 586 | 1 | 114 | Cohesin domain, interaction partner of dockerin. Bacterial cohesin domains bind to a complementary protein domain named dockerin, and this interaction is required for the formation of the cellulosome, a cellulose-degrading complex. Two specific calcium-dependent interactions between cohesin and dockerin appear to be essential for cellulosome assembly, type I and type II. This subfamily represents type III cohesins and closely related domains. |
| pfam17137 | DUF5110 | 1.95e-20 | 9 | 79 | 2 | 72 | Domain of unknown function (DUF5110). This domain is likely to be a carbohydrate-binding domain of some description as it is found immediately C-terminal to the glycosyl-hydrolase family Glyco_hydro_31, pfam01055. |
| pfam00754 | F5_F8_type_C | 7.19e-12 | 278 | 392 | 9 | 127 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| cd14254 | Dockerin_II | 8.66e-11 | 406 | 459 | 1 | 50 | Type II dockerin repeat domain. Bacterial cohesin domains bind to a complementary protein domain named dockerin, and this interaction is required for the formation of the cellulosome, a cellulose-degrading complex. The cellulosome consists of scaffoldin, a noncatalytic scaffolding polypeptide, that comprises repeating cohesion modules and a single carbohydrate-binding module (CBM). Specific calcium-dependent interactions between cohesins and dockerins appear to be essential for cellulosome assembly. This subfamily represents type II dockerins, which are responsible for mediating attachment of the cellulosome complex to the bacterial cell wall. |
| cd00057 | FA58C | 2.57e-07 | 294 | 377 | 36 | 126 | Substituted updates: Jan 31, 2002 |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QRO24224.1 | 4.62e-295 | 1 | 597 | 680 | 1276 |
| QRY59398.1 | 1.46e-241 | 6 | 599 | 694 | 1288 |
| QRQ60079.1 | 2.06e-241 | 6 | 599 | 694 | 1288 |
| QMV71333.1 | 7.64e-241 | 6 | 599 | 679 | 1273 |
| QQT54226.1 | 2.31e-240 | 6 | 599 | 694 | 1288 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6M76_A | 1.03e-37 | 6 | 263 | 702 | 963 | GH31alpha-N-acetylgalactosaminidase from Enterococcus faecalis [Enterococcus faecalis ATCC 10100],6M77_A GH31 alpha-N-acetylgalactosaminidase from Enterococcus faecalis in complex with N-acetylgalactosamine [Enterococcus faecalis ATCC 10100] |
| 7F7Q_A | 1.03e-37 | 6 | 263 | 702 | 963 | ChainA, GH31 alpha-N-acetylgalactosaminidase [Enterococcus faecalis ATCC 10100] |
| 7F7R_A | 1.03e-37 | 6 | 263 | 702 | 963 | ChainA, GH31 alpha-N-acetylgalactosaminidase [Enterococcus faecalis ATCC 10100] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as OTHER
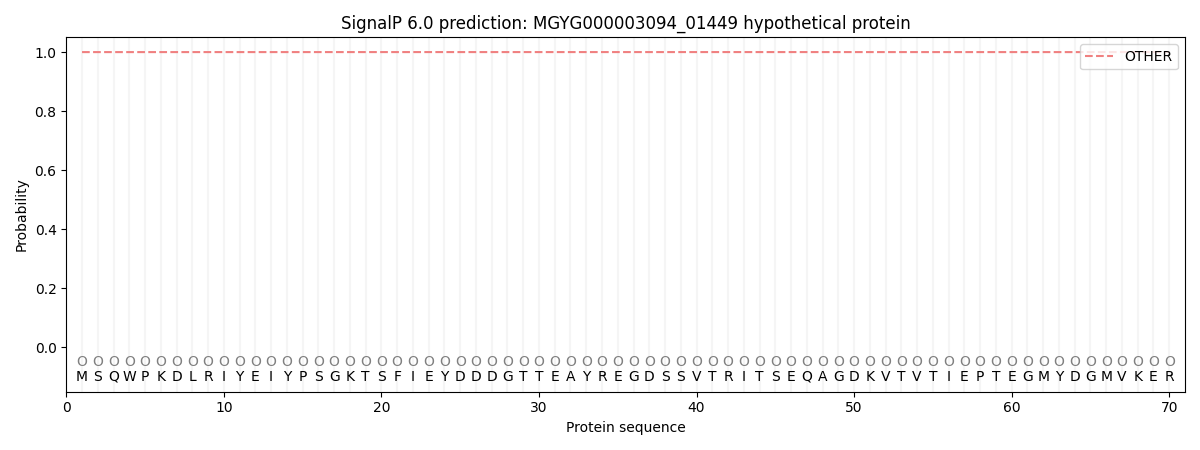
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000067 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |
