You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003095_00514
You are here: Home > Sequence: MGYG000003095_00514
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
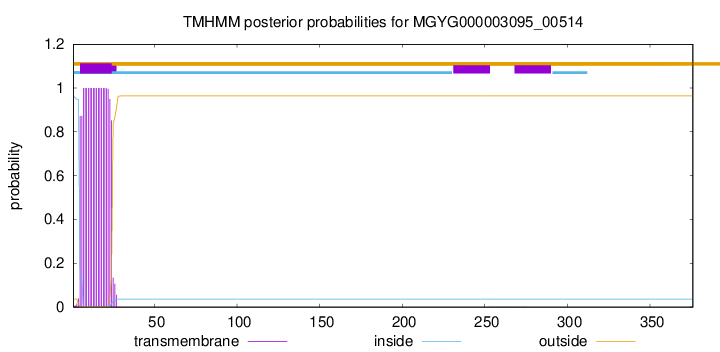
TMHMM annotations
Basic Information help
| Species | CAG-460 sp900546715 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; RF39; UBA660; CAG-460; CAG-460 sp900546715 | |||||||||||
| CAZyme ID | MGYG000003095_00514 | |||||||||||
| CAZy Family | GH3 | |||||||||||
| CAZyme Description | Beta-hexosaminidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 60267; End: 61397 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH3 | 109 | 334 | 2e-50 | 0.9768518518518519 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00933 | Glyco_hydro_3 | 2.09e-71 | 56 | 369 | 1 | 316 | Glycosyl hydrolase family 3 N terminal domain. |
| COG1472 | BglX | 5.67e-64 | 55 | 373 | 1 | 315 | Periplasmic beta-glucosidase and related glycosidases [Carbohydrate transport and metabolism]. |
| PRK05337 | PRK05337 | 6.24e-38 | 84 | 334 | 27 | 280 | beta-hexosaminidase; Provisional |
| PRK15098 | PRK15098 | 4.23e-23 | 48 | 375 | 38 | 358 | beta-glucosidase BglX. |
| cd02932 | OYE_YqiM_FMN | 0.009 | 74 | 190 | 189 | 271 | Old yellow enzyme (OYE) YqjM-like FMN binding domain. YqjM is involved in the oxidative stress response of Bacillus subtilis. Like the other OYE members, each monomer of YqjM contains FMN as a non-covalently bound cofactor and uses NADPH as a reducing agent. The YqjM enzyme exists as a homotetramer that is assembled as a dimer of catalytically dependent dimers, while other OYE members exist only as monomers or dimers. Moreover, the protein displays a shared active site architecture where an arginine finger at the COOH terminus of one monomer extends into the active site of the adjacent monomer and is directly involved in substrate recognition. Another remarkable difference in the binding of the ligand in YqjM is represented by the contribution of the NH2-terminal tyrosine instead of a COOH-terminal tyrosine in OYE and its homologs. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BCI61427.1 | 7.48e-135 | 39 | 370 | 146 | 476 |
| QHQ63470.1 | 3.82e-132 | 47 | 376 | 84 | 414 |
| QNM02560.1 | 4.16e-131 | 47 | 376 | 88 | 418 |
| ASW43722.1 | 7.18e-128 | 28 | 376 | 40 | 388 |
| BCJ98858.1 | 2.87e-126 | 47 | 376 | 127 | 457 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6K5J_A | 3.31e-45 | 55 | 370 | 11 | 337 | Structureof a glycoside hydrolase family 3 beta-N-acetylglucosaminidase from Paenibacillus sp. str. FPU-7 [Paenibacillaceae] |
| 3BMX_A | 5.03e-41 | 50 | 372 | 37 | 395 | Beta-N-hexosaminidase(YbbD) from Bacillus subtilis [Bacillus subtilis],3BMX_B Beta-N-hexosaminidase (YbbD) from Bacillus subtilis [Bacillus subtilis],3NVD_A Structure of YBBD in complex with pugnac [Bacillus subtilis],3NVD_B Structure of YBBD in complex with pugnac [Bacillus subtilis] |
| 3LK6_A | 1.96e-40 | 50 | 372 | 11 | 369 | ChainA, Lipoprotein ybbD [Bacillus subtilis],3LK6_B Chain B, Lipoprotein ybbD [Bacillus subtilis],3LK6_C Chain C, Lipoprotein ybbD [Bacillus subtilis],3LK6_D Chain D, Lipoprotein ybbD [Bacillus subtilis] |
| 4GYJ_A | 2.57e-40 | 50 | 372 | 41 | 399 | Crystalstructure of mutant (D318N) bacillus subtilis family 3 glycoside hydrolase (nagz) in complex with glcnac-murnac (space group P1) [Bacillus subtilis subsp. subtilis str. 168],4GYJ_B Crystal structure of mutant (D318N) bacillus subtilis family 3 glycoside hydrolase (nagz) in complex with glcnac-murnac (space group P1) [Bacillus subtilis subsp. subtilis str. 168],4GYK_A Crystal structure of mutant (D318N) bacillus subtilis family 3 glycoside hydrolase (nagz) in complex with glcnac-murnac (space group P1211) [Bacillus subtilis subsp. subtilis str. 168],4GYK_B Crystal structure of mutant (D318N) bacillus subtilis family 3 glycoside hydrolase (nagz) in complex with glcnac-murnac (space group P1211) [Bacillus subtilis subsp. subtilis str. 168] |
| 4YYF_A | 9.09e-34 | 59 | 355 | 43 | 340 | ChainA, Beta-N-acetylhexosaminidase [Mycolicibacterium smegmatis MC2 155],4YYF_B Chain B, Beta-N-acetylhexosaminidase [Mycolicibacterium smegmatis MC2 155],4YYF_C Chain C, Beta-N-acetylhexosaminidase [Mycolicibacterium smegmatis MC2 155] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P40406 | 2.75e-40 | 50 | 372 | 37 | 395 | Beta-hexosaminidase OS=Bacillus subtilis (strain 168) OX=224308 GN=nagZ PE=1 SV=1 |
| B2I6G9 | 5.40e-33 | 83 | 313 | 24 | 254 | Beta-hexosaminidase OS=Xylella fastidiosa (strain M23) OX=405441 GN=nagZ PE=3 SV=1 |
| Q87BR5 | 5.40e-33 | 83 | 313 | 24 | 254 | Beta-hexosaminidase OS=Xylella fastidiosa (strain Temecula1 / ATCC 700964) OX=183190 GN=nagZ PE=3 SV=1 |
| B0U3L0 | 7.50e-33 | 83 | 313 | 24 | 254 | Beta-hexosaminidase OS=Xylella fastidiosa (strain M12) OX=405440 GN=nagZ PE=3 SV=1 |
| Q9PAZ0 | 2.79e-32 | 83 | 356 | 24 | 300 | Beta-hexosaminidase OS=Xylella fastidiosa (strain 9a5c) OX=160492 GN=nagZ PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.749500 | 0.232246 | 0.013131 | 0.001028 | 0.000691 | 0.003405 |