You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003104_00116
You are here: Home > Sequence: MGYG000003104_00116
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
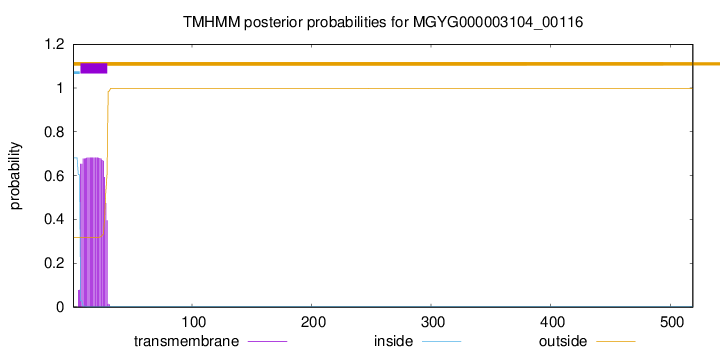
TMHMM annotations
Basic Information help
| Species | UMGS1601 sp900545345 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Ruminococcaceae; UMGS1601; UMGS1601 sp900545345 | |||||||||||
| CAZyme ID | MGYG000003104_00116 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 127817; End: 129376 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 81 | 354 | 4.3e-99 | 0.9891304347826086 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00150 | Cellulase | 3.66e-56 | 67 | 355 | 2 | 270 | Cellulase (glycosyl hydrolase family 5). |
| COG2730 | BglC | 3.64e-33 | 1 | 364 | 1 | 370 | Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
| COG1388 | LysM | 0.009 | 430 | 503 | 3 | 76 | LysM repeat [Cell wall/membrane/envelope biogenesis]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| CBL35063.1 | 2.87e-173 | 34 | 374 | 138 | 482 |
| CBK96026.1 | 3.99e-172 | 34 | 374 | 93 | 437 |
| CCO05659.1 | 9.72e-155 | 2 | 373 | 3 | 383 |
| CBL17363.1 | 1.72e-95 | 15 | 374 | 9 | 371 |
| QQA02033.1 | 3.15e-86 | 44 | 359 | 47 | 357 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6WQP_A | 1.43e-64 | 39 | 373 | 13 | 352 | GH5-4broad specificity endoglucanase from Ruminococcus champanellensis [Ruminococcus champanellensis],6WQP_B GH5-4 broad specificity endoglucanase from Ruminococcus champanellensis [Ruminococcus champanellensis],6WQV_A GH5-4 broad specificity endoglucanase from Ruminococcus champanellensis with bound cellotriose [Ruminococcus champanellensis],6WQV_B GH5-4 broad specificity endoglucanase from Ruminococcus champanellensis with bound cellotriose [Ruminococcus champanellensis],6WQV_C GH5-4 broad specificity endoglucanase from Ruminococcus champanellensis with bound cellotriose [Ruminococcus champanellensis],6WQV_D GH5-4 broad specificity endoglucanase from Ruminococcus champanellensis with bound cellotriose [Ruminococcus champanellensis] |
| 4YZP_A | 4.59e-64 | 41 | 367 | 23 | 321 | Crystalstructure of a tri-modular GH5 (subfamily 4) endo-beta-1, 4-glucanase from Bacillus licheniformis [Bacillus licheniformis],4YZT_A Crystal structure of a tri-modular GH5 (subfamily 4) endo-beta-1, 4-glucanase from Bacillus licheniformis complexed with cellotetraose [Bacillus licheniformis] |
| 2JEP_A | 2.37e-63 | 5 | 376 | 6 | 393 | Nativefamily 5 xyloglucanase from Paenibacillus pabuli [Paenibacillus pabuli],2JEP_B Native family 5 xyloglucanase from Paenibacillus pabuli [Paenibacillus pabuli],2JEQ_A Family 5 xyloglucanase from Paenibacillus pabuli in complex with ligand [Paenibacillus pabuli] |
| 4IM4_A | 3.58e-63 | 39 | 376 | 4 | 334 | ChainA, Endoglucanase E [Acetivibrio thermocellus],4IM4_B Chain B, Endoglucanase E [Acetivibrio thermocellus],4IM4_C Chain C, Endoglucanase E [Acetivibrio thermocellus],4IM4_D Chain D, Endoglucanase E [Acetivibrio thermocellus],4IM4_E Chain E, Endoglucanase E [Acetivibrio thermocellus],4IM4_F Chain F, Endoglucanase E [Acetivibrio thermocellus] |
| 6WQY_A | 1.38e-62 | 41 | 364 | 23 | 357 | ChainA, Cellulase [Phocaeicola salanitronis DSM 18170] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q12647 | 5.03e-60 | 22 | 376 | 6 | 362 | Endoglucanase B OS=Neocallimastix patriciarum OX=4758 GN=CELB PE=2 SV=1 |
| O08342 | 1.73e-59 | 41 | 354 | 39 | 369 | Endoglucanase A OS=Paenibacillus barcinonensis OX=198119 GN=celA PE=1 SV=1 |
| P10477 | 1.49e-58 | 39 | 376 | 54 | 384 | Cellulase/esterase CelE OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=celE PE=1 SV=2 |
| P28621 | 3.48e-58 | 39 | 376 | 39 | 373 | Endoglucanase B OS=Clostridium cellulovorans (strain ATCC 35296 / DSM 3052 / OCM 3 / 743B) OX=573061 GN=engB PE=3 SV=1 |
| P23660 | 2.05e-57 | 28 | 373 | 14 | 359 | Endoglucanase A OS=Ruminococcus albus OX=1264 GN=celA PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
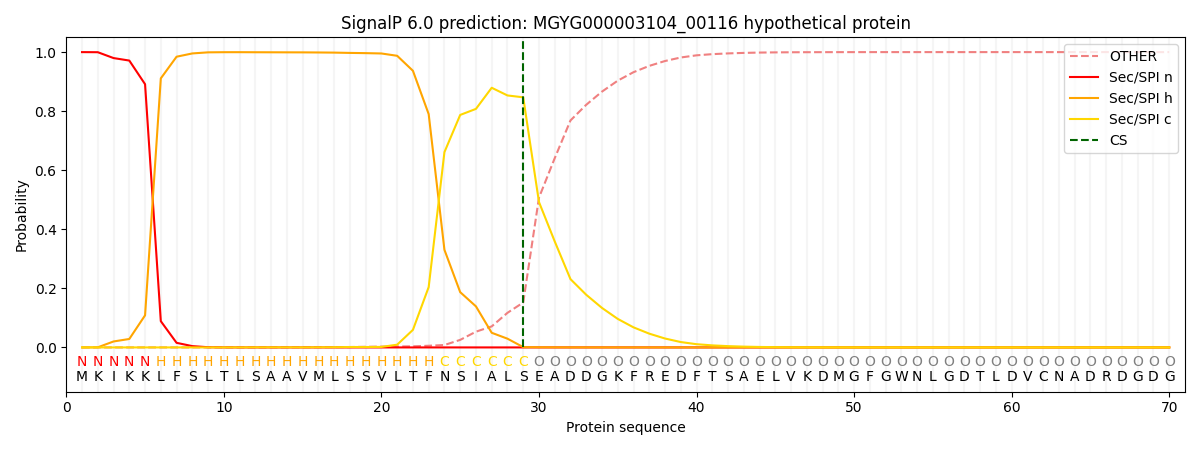
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000397 | 0.998814 | 0.000244 | 0.000188 | 0.000163 | 0.000159 |