You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003114_02422
You are here: Home > Sequence: MGYG000003114_02422
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
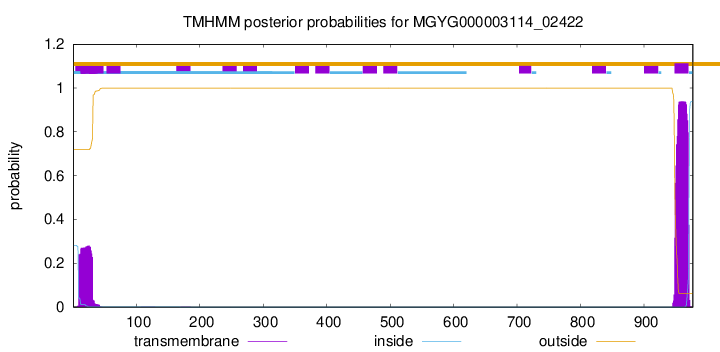
TMHMM annotations
Basic Information help
| Species | Clostridium sp900547475 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Clostridiales; Clostridiaceae; Clostridium; Clostridium sp900547475 | |||||||||||
| CAZyme ID | MGYG000003114_02422 | |||||||||||
| CAZy Family | CBM27 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 17833; End: 20766 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH26 | 237 | 566 | 2.3e-60 | 0.9141914191419142 |
| CBM23 | 721 | 890 | 2.5e-39 | 0.9814814814814815 |
| CBM27 | 43 | 200 | 1.4e-25 | 0.9821428571428571 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam02156 | Glyco_hydro_26 | 7.88e-44 | 237 | 491 | 1 | 242 | Glycosyl hydrolase family 26. |
| COG4124 | ManB2 | 4.77e-21 | 368 | 574 | 145 | 352 | Beta-mannanase [Carbohydrate transport and metabolism]. |
| pfam03425 | CBM_11 | 2.29e-10 | 717 | 845 | 3 | 128 | Carbohydrate binding domain (family 11). |
| pfam08547 | CIA30 | 4.72e-07 | 752 | 863 | 23 | 142 | Complex I intermediate-associated protein 30 (CIA30). This protein is associated with mitochondrial Complex I intermediate-associated protein 30 (CIA30) in human and mouse. The family is also present in Schizosaccharomyces pombe which does not contain the NADH dehydrogenase component of complex I, or many of the other essential subunits. This means it is possible that this family of protein may not be directly involved in oxidative phosphorylation. |
| pfam00746 | Gram_pos_anchor | 4.57e-05 | 938 | 977 | 2 | 41 | LPXTG cell wall anchor motif. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUF84291.1 | 0.0 | 36 | 898 | 275 | 1133 |
| AXB85606.1 | 0.0 | 31 | 898 | 93 | 956 |
| QSX03803.1 | 0.0 | 36 | 898 | 275 | 1133 |
| QJU44978.1 | 0.0 | 36 | 898 | 275 | 1133 |
| QCJ07246.1 | 0.0 | 36 | 898 | 275 | 1133 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4YN5_A | 7.88e-66 | 232 | 600 | 47 | 414 | Catalyticdomain of Bacillus sp. JAMB-750 GH26 Endo-beta-1,4-mannanase [Bacillus sp. JAMB750] |
| 1J9Y_A | 4.64e-56 | 231 | 600 | 6 | 369 | Crystalstructure of mannanase 26A from Pseudomonas cellulosa [Cellvibrio japonicus] |
| 2BVT_A | 4.68e-56 | 232 | 642 | 4 | 397 | Thestructure of a modular endo-beta-1,4-mannanase from Cellulomonas fimi explains the product specificity of glycoside hydrolase family 26 mannanases. [Cellulomonas fimi],2BVT_B The structure of a modular endo-beta-1,4-mannanase from Cellulomonas fimi explains the product specificity of glycoside hydrolase family 26 mannanases. [Cellulomonas fimi],2BVY_A The structure and characterization of a modular endo-beta-1,4-mannanase from Cellulomonas fimi [Cellulomonas fimi] |
| 1R7O_A | 6.00e-56 | 231 | 600 | 16 | 379 | CrystalStructure of apo-mannanase 26A from Psudomonas cellulosa [Cellvibrio japonicus] |
| 1GW1_A | 9.37e-56 | 231 | 600 | 2 | 362 | Substratedistortion by beta-mannanase from Pseudomonas cellulosa [Cellvibrio japonicus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P49424 | 6.62e-55 | 231 | 600 | 44 | 407 | Mannan endo-1,4-beta-mannosidase OS=Cellvibrio japonicus (strain Ueda107) OX=498211 GN=manA PE=1 SV=2 |
| A1A278 | 6.94e-55 | 233 | 853 | 39 | 649 | Mannan endo-1,4-beta-mannosidase OS=Bifidobacterium adolescentis (strain ATCC 15703 / DSM 20083 / NCTC 11814 / E194a) OX=367928 GN=BAD_1030 PE=1 SV=1 |
| P16699 | 1.50e-21 | 221 | 483 | 18 | 274 | Mannan endo-1,4-beta-mannosidase A and B OS=Caldalkalibacillus mannanilyticus (strain DSM 16130 / CIP 109019 / JCM 10596 / AM-001) OX=1236954 PE=1 SV=1 |
| Q5AWB7 | 1.07e-18 | 294 | 517 | 76 | 298 | Probable mannan endo-1,4-beta-mannosidase E OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=manE PE=3 SV=1 |
| P55278 | 3.03e-16 | 325 | 483 | 121 | 270 | Mannan endo-1,4-beta-mannosidase OS=Bacillus subtilis OX=1423 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
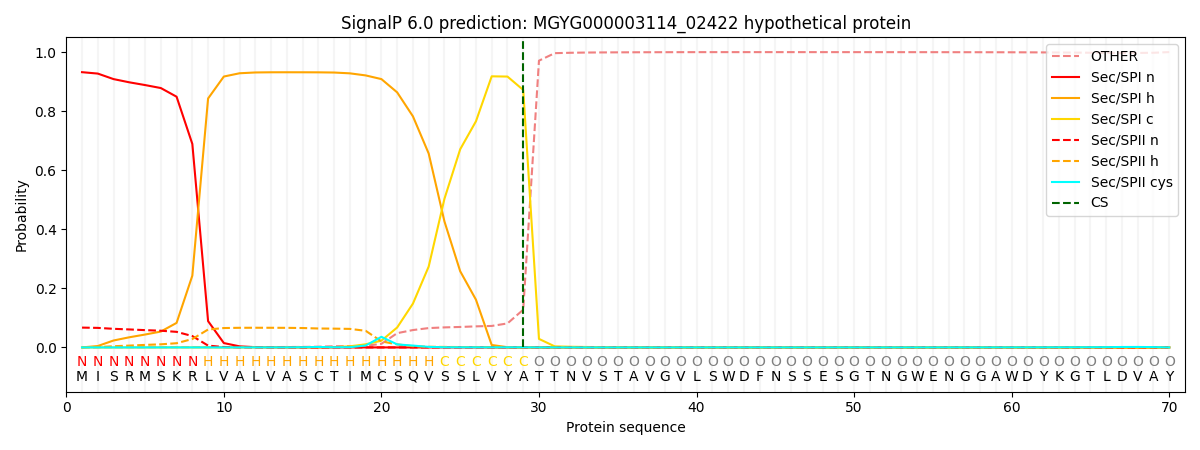
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.002911 | 0.922824 | 0.072755 | 0.000594 | 0.000458 | 0.000425 |