You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003116_00864
You are here: Home > Sequence: MGYG000003116_00864
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
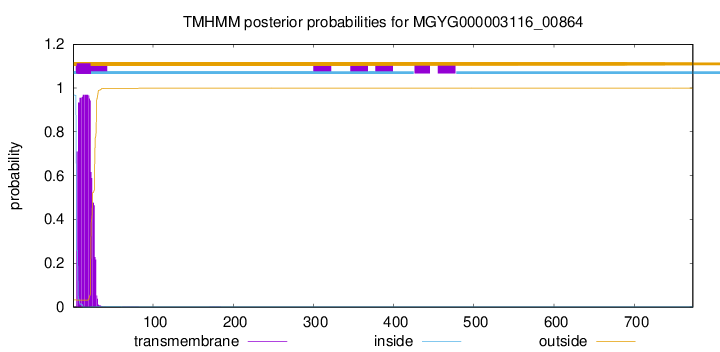
TMHMM annotations
Basic Information help
| Species | Clostridium_F tepidum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Clostridiales; Clostridiaceae; Clostridium_F; Clostridium_F tepidum | |||||||||||
| CAZyme ID | MGYG000003116_00864 | |||||||||||
| CAZy Family | CBM5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 137400; End: 139721 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam11958 | DUF3472 | 9.39e-40 | 92 | 249 | 3 | 173 | Domain of unknown function (DUF3472). This presumed domain is functionally uncharacterized. This domain is found in bacteria, eukaryotes and viruses. This domain is typically between 174 to 190 amino acids in length. This domain has a single completely conserved residue G that may be functionally important. |
| pfam03272 | Mucin_bdg | 2.19e-32 | 380 | 494 | 1 | 116 | Putative mucin or carbohydrate-binding module. This family is the putative binding domain for the substrates of enhancin, and other similar metallopeptidases. This is not the enzymically active, peptidase, part of the proteins - see pfam13402. |
| pfam03272 | Mucin_bdg | 5.30e-27 | 518 | 632 | 1 | 116 | Putative mucin or carbohydrate-binding module. This family is the putative binding domain for the substrates of enhancin, and other similar metallopeptidases. This is not the enzymically active, peptidase, part of the proteins - see pfam13402. |
| pfam03272 | Mucin_bdg | 1.57e-17 | 657 | 769 | 2 | 116 | Putative mucin or carbohydrate-binding module. This family is the putative binding domain for the substrates of enhancin, and other similar metallopeptidases. This is not the enzymically active, peptidase, part of the proteins - see pfam13402. |
| TIGR01076 | sortase_fam | 2.90e-04 | 639 | 746 | 4 | 109 | LPXTG-site transpeptidase (sortase) family protein. This family includes Staphylococcus aureus sortase, a transpeptidase that attaches surface proteins by the Thr of an LPXTG motif to the cell wall. It also includes a protein required for correct assembly of an LPXTG-containing fimbrial protein, a set of homologous proteins from Streptococcus pneumoniae, in which LPXTG proteins are common. However, related proteins are found in Bacillus subtilis and Methanobacterium thermoautotrophicum, in which LPXTG-mediated cell wall attachment is not known. [Cell envelope, Other, Protein fate, Protein and peptide secretion and trafficking] |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AUM88102.1 | 0.0 | 1 | 637 | 1 | 637 |
| ACA54402.1 | 0.0 | 1 | 645 | 1 | 652 |
| AUN10863.1 | 0.0 | 1 | 637 | 1 | 637 |
| APH19613.1 | 0.0 | 1 | 640 | 1 | 640 |
| AUM91712.1 | 0.0 | 1 | 640 | 1 | 640 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
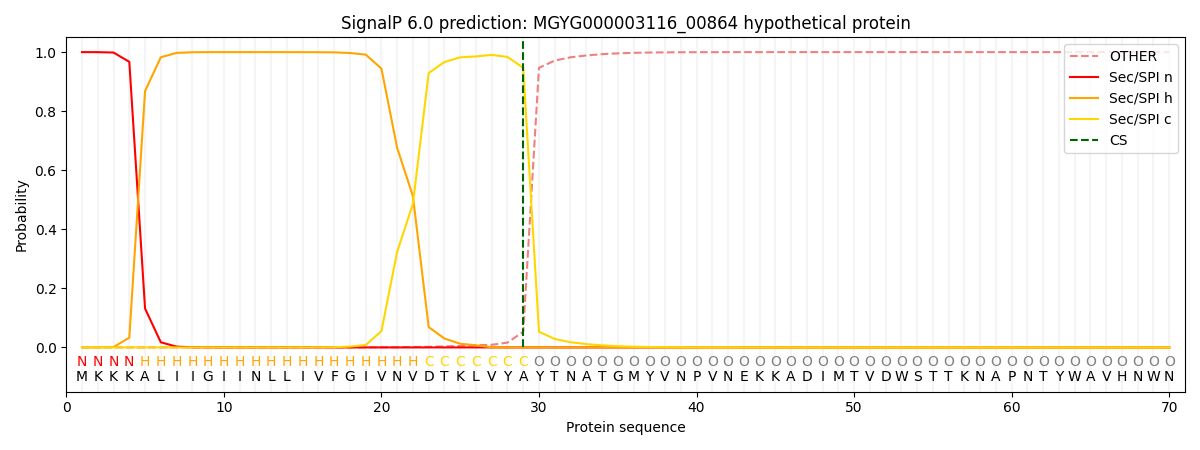
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.001153 | 0.997950 | 0.000288 | 0.000238 | 0.000173 | 0.000170 |