You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003123_03190
You are here: Home > Sequence: MGYG000003123_03190
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Chimaeribacter coloradensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Proteobacteria; Gammaproteobacteria; Enterobacterales; Enterobacteriaceae; Chimaeribacter; Chimaeribacter coloradensis | |||||||||||
| CAZyme ID | MGYG000003123_03190 | |||||||||||
| CAZy Family | GH23 | |||||||||||
| CAZyme Description | Endo-type membrane-bound lytic murein transglycosylase A | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 3554; End: 4186 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH23 | 56 | 205 | 3.8e-21 | 0.8074074074074075 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd16893 | LT_MltC_MltE | 1.50e-86 | 47 | 204 | 3 | 162 | membrane-bound lytic murein transglycosylases MltC and MltE, and similar proteins. MltC and MltE are periplasmic, outer membrane attached lytic transglycosylases (LTs), which cleave beta-1,4-glycosidic bonds joining N-acetylmuramic acid and N-acetylglucosamine in the cell wall peptidoglycan, yielding 1,6-anhydromuropeptides. Proteins similar to this family include the soluble and insoluble membrane-bound LTs in bacteria and the LTs in bacteriophage lambda |
| PRK15470 | emtA | 6.04e-74 | 49 | 206 | 45 | 203 | membrane-bound lytic murein transglycosylase EmtA. |
| PRK11671 | mltC | 8.55e-68 | 47 | 205 | 196 | 355 | membrane-bound lytic murein transglycosylase MltC. |
| COG0741 | MltE | 2.17e-33 | 39 | 202 | 135 | 294 | Soluble lytic murein transglycosylase and related regulatory proteins (some contain LysM/invasin domains) [Cell wall/membrane/envelope biogenesis]. |
| pfam01464 | SLT | 2.33e-31 | 47 | 171 | 1 | 114 | Transglycosylase SLT domain. This family is distantly related to pfam00062. Members are found in phages, type II, type III and type IV secretion systems. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QCR36319.1 | 7.53e-115 | 3 | 210 | 1 | 212 |
| QMV51345.1 | 3.91e-91 | 9 | 206 | 7 | 209 |
| ANI31905.1 | 1.03e-90 | 3 | 206 | 1 | 216 |
| ATA22603.1 | 9.99e-90 | 3 | 206 | 1 | 219 |
| QQX53346.1 | 3.59e-87 | 3 | 208 | 1 | 221 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2Y8P_A | 7.52e-60 | 38 | 207 | 17 | 187 | CrystalStructure of an Outer Membrane-Anchored Endolytic Peptidoglycan Lytic Transglycosylase (MltE) from Escherichia coli [Escherichia coli K-12],2Y8P_B Crystal Structure of an Outer Membrane-Anchored Endolytic Peptidoglycan Lytic Transglycosylase (MltE) from Escherichia coli [Escherichia coli K-12] |
| 6GHY_A | 2.14e-59 | 38 | 207 | 17 | 187 | Structureof Lytic Transglycosylase MltE inactive mutant E64Q from E.coli [Escherichia coli K-12],6GHY_B Structure of Lytic Transglycosylase MltE inactive mutant E64Q from E.coli [Escherichia coli K-12] |
| 6GI3_B | 2.14e-59 | 38 | 207 | 17 | 187 | Structureof Lytic Transglycosylase MltE mutant S73A from E.coli [Escherichia coli] |
| 6GI4_B | 2.14e-59 | 38 | 207 | 17 | 187 | Structureof Lytic Transglycosylase MltE mutant S75A from E.coli [Escherichia coli] |
| 6GHZ_A | 3.04e-59 | 38 | 207 | 17 | 187 | Structureof Lytic Transglycosylase MltE mutant Y192F from E.coli [Escherichia coli K-12],6GHZ_B Structure of Lytic Transglycosylase MltE mutant Y192F from E.coli [Escherichia coli K-12] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| A4WBE8 | 3.35e-65 | 9 | 206 | 6 | 203 | Endo-type membrane-bound lytic murein transglycosylase A OS=Enterobacter sp. (strain 638) OX=399742 GN=emtA PE=3 SV=1 |
| A7MKC3 | 1.56e-63 | 6 | 206 | 2 | 203 | Endo-type membrane-bound lytic murein transglycosylase A OS=Cronobacter sakazakii (strain ATCC BAA-894) OX=290339 GN=emtA PE=3 SV=2 |
| A8AFS5 | 2.21e-63 | 9 | 206 | 6 | 203 | Endo-type membrane-bound lytic murein transglycosylase A OS=Citrobacter koseri (strain ATCC BAA-895 / CDC 4225-83 / SGSC4696) OX=290338 GN=emtA PE=3 SV=1 |
| Q8XGT6 | 4.44e-63 | 9 | 206 | 6 | 203 | Endo-type membrane-bound lytic murein transglycosylase A OS=Salmonella typhi OX=90370 GN=emtA PE=3 SV=1 |
| Q57NL3 | 4.44e-63 | 9 | 206 | 6 | 203 | Endo-type membrane-bound lytic murein transglycosylase A OS=Salmonella choleraesuis (strain SC-B67) OX=321314 GN=emtA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
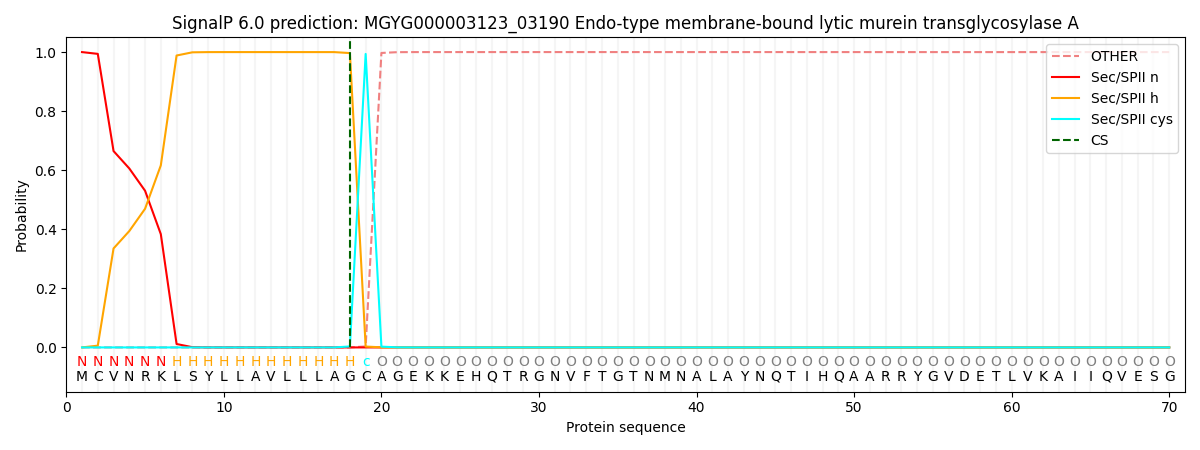
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000000 | 1.000048 | 0.000000 | 0.000000 | 0.000000 |
