You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003125_01195
You are here: Home > Sequence: MGYG000003125_01195
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Corynebacterium kefirresidentii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Mycobacteriales; Mycobacteriaceae; Corynebacterium; Corynebacterium kefirresidentii | |||||||||||
| CAZyme ID | MGYG000003125_01195 | |||||||||||
| CAZy Family | CE1 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 58589; End: 59548 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE1 | 69 | 224 | 4.6e-27 | 0.5947136563876652 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3509 | LpqC | 4.24e-40 | 52 | 312 | 32 | 309 | Poly(3-hydroxybutyrate) depolymerase [Secondary metabolites biosynthesis, transport and catabolism]. |
| COG0400 | YpfH | 6.78e-11 | 80 | 225 | 13 | 167 | Predicted esterase [General function prediction only]. |
| COG4099 | COG4099 | 5.18e-10 | 71 | 189 | 176 | 301 | Predicted peptidase [General function prediction only]. |
| pfam10503 | Esterase_phd | 6.87e-08 | 86 | 209 | 17 | 144 | Esterase PHB depolymerase. This family of proteins include acetyl xylan esterases (AXE), feruloyl esterases (FAE), and poly(3-hydroxybutyrate) (PHB) depolymerases. |
| COG1506 | DAP2 | 1.48e-07 | 35 | 193 | 345 | 508 | Dipeptidyl aminopeptidase/acylaminoacyl peptidase [Amino acid transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ALO15718.1 | 1.16e-21 | 69 | 317 | 35 | 299 |
| AUX40426.1 | 2.40e-19 | 39 | 307 | 226 | 463 |
| AGP34778.1 | 3.36e-19 | 39 | 307 | 233 | 470 |
| BAJ10857.1 | 9.83e-19 | 46 | 249 | 19 | 207 |
| AUX29931.1 | 1.12e-18 | 42 | 307 | 233 | 467 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| A1CC33 | 5.35e-18 | 54 | 307 | 30 | 269 | Probable feruloyl esterase C OS=Aspergillus clavatus (strain ATCC 1007 / CBS 513.65 / DSM 816 / NCTC 3887 / NRRL 1 / QM 1276 / 107) OX=344612 GN=faeC-2 PE=3 SV=1 |
| Q9Y871 | 3.31e-12 | 69 | 247 | 292 | 484 | Feruloyl esterase B OS=Piromyces equi OX=99929 GN=ESTA PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
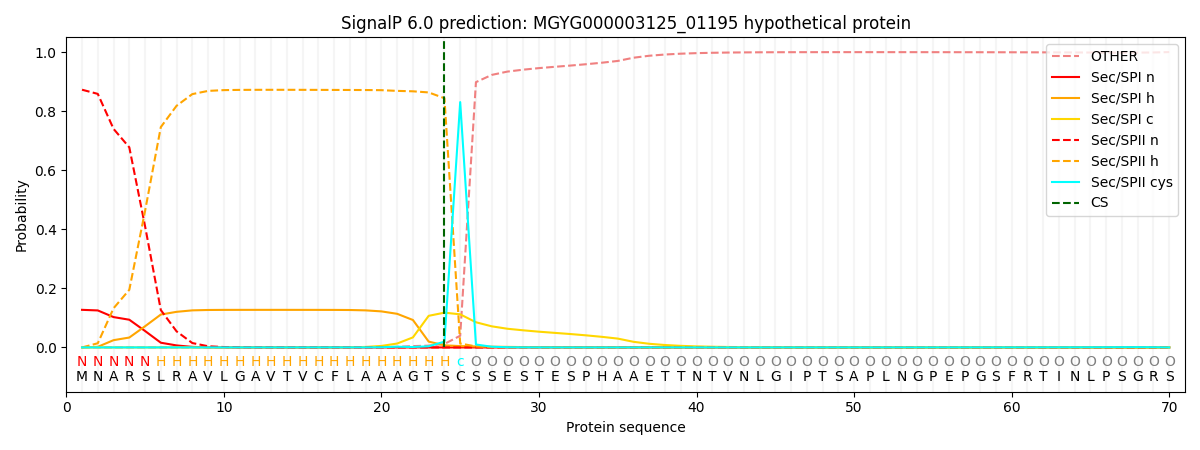
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000541 | 0.122617 | 0.876238 | 0.000244 | 0.000206 | 0.000133 |
