You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003132_01163
You are here: Home > Sequence: MGYG000003132_01163
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Dysgonomonas sp900556485 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Dysgonomonadaceae; Dysgonomonas; Dysgonomonas sp900556485 | |||||||||||
| CAZyme ID | MGYG000003132_01163 | |||||||||||
| CAZy Family | GH9 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 115648; End: 118092 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH9 | 335 | 760 | 1.8e-34 | 0.8444976076555024 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd02850 | E_set_Cellulase_N | 3.64e-15 | 245 | 327 | 2 | 86 | N-terminal Early set domain associated with the catalytic domain of cellulase. E or "early" set domains are associated with the catalytic domain of cellulases at the N-terminal end. Cellulases are O-glycosyl hydrolases (GHs) that hydrolyze beta 1-4 glucosidic bonds in cellulose. They are usually categorized into either exoglucanases, which sequentially release terminal sugar units from the cellulose chain, or endoglucanases, which also attack the chain internally. The N-terminal domain of cellulase may be related to the immunoglobulin and/or fibronectin type III superfamilies. These domains are associated with different types of catalytic domains at either the N-terminal or C-terminal end and may be involved in homodimeric/tetrameric/dodecameric interactions. Members of this family include members of the alpha amylase family, sialidase, galactose oxidase, cellulase, cellulose, hyaluronate lyase, chitobiase, and chitinase, among others. |
| pfam02927 | CelD_N | 2.41e-11 | 251 | 321 | 9 | 82 | Cellulase N-terminal ig-like domain. |
| pfam00759 | Glyco_hydro_9 | 3.43e-09 | 340 | 558 | 7 | 238 | Glycosyl hydrolase family 9. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QMU26859.1 | 4.32e-22 | 251 | 813 | 71 | 618 |
| ARW20213.1 | 9.00e-21 | 750 | 813 | 1 | 64 |
| AFY92150.1 | 1.03e-20 | 235 | 435 | 24 | 249 |
| AFN75471.1 | 1.66e-19 | 251 | 434 | 36 | 233 |
| AFZ21278.1 | 1.98e-18 | 241 | 498 | 87 | 371 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5DGQ_A | 9.39e-08 | 296 | 436 | 58 | 210 | Crystalstructure of GH9 exo-beta-D-glucosaminidase PBPRA0520 [Photobacterium profundum],5DGQ_B Crystal structure of GH9 exo-beta-D-glucosaminidase PBPRA0520 [Photobacterium profundum],5DGR_A Crystal structure of GH9 exo-beta-D-glucosaminidase PBPRA0520, glucosamine complex [Photobacterium profundum],5DGR_B Crystal structure of GH9 exo-beta-D-glucosaminidase PBPRA0520, glucosamine complex [Photobacterium profundum] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
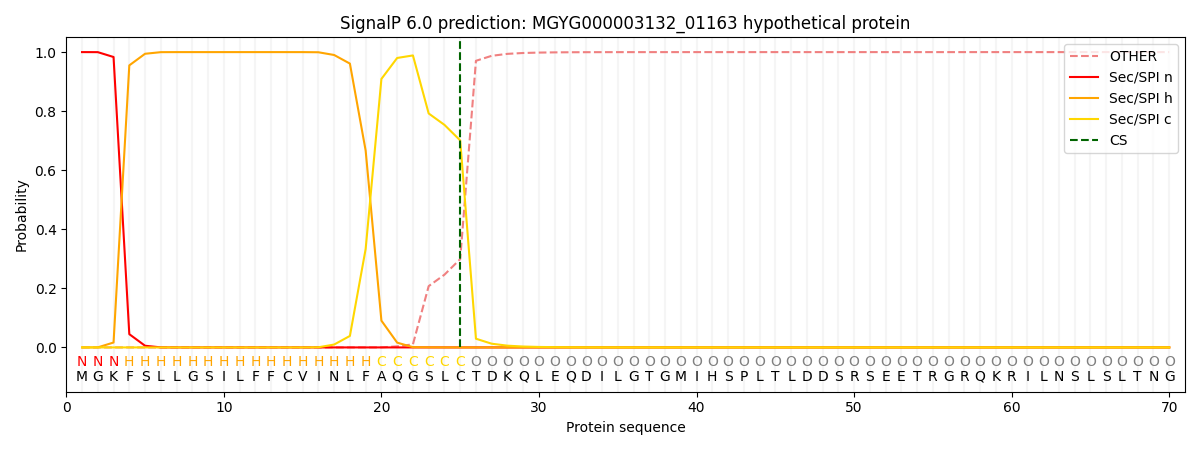
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000243 | 0.999189 | 0.000153 | 0.000140 | 0.000127 | 0.000125 |
