You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003132_02099
You are here: Home > Sequence: MGYG000003132_02099
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Dysgonomonas sp900556485 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Dysgonomonadaceae; Dysgonomonas; Dysgonomonas sp900556485 | |||||||||||
| CAZyme ID | MGYG000003132_02099 | |||||||||||
| CAZy Family | CE1 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 57314; End: 58072 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE1 | 31 | 234 | 7.3e-31 | 0.8898678414096917 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG4099 | COG4099 | 6.22e-49 | 29 | 252 | 171 | 387 | Predicted peptidase [General function prediction only]. |
| COG1506 | DAP2 | 3.70e-19 | 29 | 250 | 375 | 616 | Dipeptidyl aminopeptidase/acylaminoacyl peptidase [Amino acid transport and metabolism]. |
| COG0400 | YpfH | 1.06e-11 | 49 | 247 | 19 | 202 | Predicted esterase [General function prediction only]. |
| pfam02230 | Abhydrolase_2 | 5.71e-11 | 49 | 234 | 15 | 202 | Phospholipase/Carboxylesterase. This family consists of both phospholipases and carboxylesterases with broad substrate specificity, and is structurally related to alpha/beta hydrolases pfam00561. |
| COG1073 | FrsA | 1.12e-06 | 29 | 250 | 30 | 297 | Fermentation-respiration switch protein FrsA, has esterase activity, DUF1100 family [Signal transduction mechanisms]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BCI61582.1 | 9.76e-47 | 32 | 248 | 825 | 1039 |
| ABS60377.1 | 9.94e-45 | 32 | 252 | 22 | 245 |
| QDU56037.1 | 1.54e-35 | 32 | 252 | 801 | 1007 |
| ACR12533.1 | 3.10e-28 | 31 | 249 | 63 | 278 |
| VTR91196.1 | 5.01e-28 | 32 | 252 | 43 | 239 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3DOH_A | 1.71e-49 | 29 | 252 | 155 | 380 | CrystalStructure of a Thermostable Esterase [Thermotoga maritima],3DOH_B Crystal Structure of a Thermostable Esterase [Thermotoga maritima],3DOI_A Crystal Structure of a Thermostable Esterase complex with paraoxon [Thermotoga maritima],3DOI_B Crystal Structure of a Thermostable Esterase complex with paraoxon [Thermotoga maritima] |
| 3WYD_A | 1.74e-24 | 28 | 252 | 17 | 217 | C-terminalesterase domain of LC-Est1 [uncultured organism],3WYD_B C-terminal esterase domain of LC-Est1 [uncultured organism] |
| 5DWD_A | 5.63e-07 | 50 | 231 | 43 | 222 | Crystalstructure of esterase PE8 [Pelagibacterium halotolerans B2],5DWD_B Crystal structure of esterase PE8 [Pelagibacterium halotolerans B2] |
| 1ORV_A | 9.02e-06 | 34 | 237 | 488 | 708 | CrystalStructure of Porcine Dipeptidyl Peptidase IV (CD26) [Sus scrofa],1ORV_B Crystal Structure of Porcine Dipeptidyl Peptidase IV (CD26) [Sus scrofa],1ORV_C Crystal Structure of Porcine Dipeptidyl Peptidase IV (CD26) [Sus scrofa],1ORV_D Crystal Structure of Porcine Dipeptidyl Peptidase IV (CD26) [Sus scrofa],1ORW_A Crystal Structure of Porcine Dipeptidyl Peptidase IV (CD26) in Complex with a Peptidomimetic Inhibitor [Sus scrofa],1ORW_B Crystal Structure of Porcine Dipeptidyl Peptidase IV (CD26) in Complex with a Peptidomimetic Inhibitor [Sus scrofa],1ORW_C Crystal Structure of Porcine Dipeptidyl Peptidase IV (CD26) in Complex with a Peptidomimetic Inhibitor [Sus scrofa],1ORW_D Crystal Structure of Porcine Dipeptidyl Peptidase IV (CD26) in Complex with a Peptidomimetic Inhibitor [Sus scrofa],2AJ8_A Porcine dipeptidyl peptidase IV (CD26) in complex with 7-Benzyl-1,3-dimethyl-8-piperazin-1-yl-3,7-dihydro-purine-2,6-dione (BDPX) [Sus scrofa],2AJ8_B Porcine dipeptidyl peptidase IV (CD26) in complex with 7-Benzyl-1,3-dimethyl-8-piperazin-1-yl-3,7-dihydro-purine-2,6-dione (BDPX) [Sus scrofa],2AJ8_C Porcine dipeptidyl peptidase IV (CD26) in complex with 7-Benzyl-1,3-dimethyl-8-piperazin-1-yl-3,7-dihydro-purine-2,6-dione (BDPX) [Sus scrofa],2AJ8_D Porcine dipeptidyl peptidase IV (CD26) in complex with 7-Benzyl-1,3-dimethyl-8-piperazin-1-yl-3,7-dihydro-purine-2,6-dione (BDPX) [Sus scrofa],2AJB_A Porcine dipeptidyl peptidase IV (CD26) in complex with the tripeptide tert-butyl-Gly-L-Pro-L-Ile (tBu-GPI) [Sus scrofa],2AJB_B Porcine dipeptidyl peptidase IV (CD26) in complex with the tripeptide tert-butyl-Gly-L-Pro-L-Ile (tBu-GPI) [Sus scrofa],2AJB_C Porcine dipeptidyl peptidase IV (CD26) in complex with the tripeptide tert-butyl-Gly-L-Pro-L-Ile (tBu-GPI) [Sus scrofa],2AJB_D Porcine dipeptidyl peptidase IV (CD26) in complex with the tripeptide tert-butyl-Gly-L-Pro-L-Ile (tBu-GPI) [Sus scrofa],2AJC_A Porcine dipeptidyl peptidase IV (CD26) in complex with 4-(2-Aminoethyl)-benzene sulphonyl fluoride (AEBSF) [Sus scrofa],2AJC_B Porcine dipeptidyl peptidase IV (CD26) in complex with 4-(2-Aminoethyl)-benzene sulphonyl fluoride (AEBSF) [Sus scrofa],2AJC_C Porcine dipeptidyl peptidase IV (CD26) in complex with 4-(2-Aminoethyl)-benzene sulphonyl fluoride (AEBSF) [Sus scrofa],2AJC_D Porcine dipeptidyl peptidase IV (CD26) in complex with 4-(2-Aminoethyl)-benzene sulphonyl fluoride (AEBSF) [Sus scrofa],2AJD_A Porcine dipeptidyl peptidase IV (CD26) in complex with L-Pro-boro-L-Pro (boroPro) [Sus scrofa],2AJD_B Porcine dipeptidyl peptidase IV (CD26) in complex with L-Pro-boro-L-Pro (boroPro) [Sus scrofa],2AJD_C Porcine dipeptidyl peptidase IV (CD26) in complex with L-Pro-boro-L-Pro (boroPro) [Sus scrofa],2AJD_D Porcine dipeptidyl peptidase IV (CD26) in complex with L-Pro-boro-L-Pro (boroPro) [Sus scrofa],2BUA_A Crystal Structure Of Porcine Dipeptidyl Peptidase IV (Cd26) in Complex With a Low Molecular Weight Inhibitor. [Sus scrofa],2BUA_B Crystal Structure Of Porcine Dipeptidyl Peptidase IV (Cd26) in Complex With a Low Molecular Weight Inhibitor. [Sus scrofa],2BUA_C Crystal Structure Of Porcine Dipeptidyl Peptidase IV (Cd26) in Complex With a Low Molecular Weight Inhibitor. [Sus scrofa],2BUA_D Crystal Structure Of Porcine Dipeptidyl Peptidase IV (Cd26) in Complex With a Low Molecular Weight Inhibitor. [Sus scrofa],2BUC_A Crystal Structure Of Porcine Dipeptidyl Peptidase IV (CD26) in Complex with a Tetrahydroisoquinoline Inhibitor [Sus scrofa],2BUC_B Crystal Structure Of Porcine Dipeptidyl Peptidase IV (CD26) in Complex with a Tetrahydroisoquinoline Inhibitor [Sus scrofa],2BUC_C Crystal Structure Of Porcine Dipeptidyl Peptidase IV (CD26) in Complex with a Tetrahydroisoquinoline Inhibitor [Sus scrofa],2BUC_D Crystal Structure Of Porcine Dipeptidyl Peptidase IV (CD26) in Complex with a Tetrahydroisoquinoline Inhibitor [Sus scrofa] |
| 5LLS_A | 9.13e-06 | 34 | 237 | 526 | 746 | Porcinedipeptidyl peptidase IV in complex with 8-(3-aminopiperidin-1-yl)-7-[(2-bromophenyl)methyl]-1,3-dimethyl-2,3,6,7-tetrahydro-1H-purine-2,6-dione [Sus scrofa],5LLS_B Porcine dipeptidyl peptidase IV in complex with 8-(3-aminopiperidin-1-yl)-7-[(2-bromophenyl)methyl]-1,3-dimethyl-2,3,6,7-tetrahydro-1H-purine-2,6-dione [Sus scrofa],5LLS_C Porcine dipeptidyl peptidase IV in complex with 8-(3-aminopiperidin-1-yl)-7-[(2-bromophenyl)methyl]-1,3-dimethyl-2,3,6,7-tetrahydro-1H-purine-2,6-dione [Sus scrofa],5LLS_D Porcine dipeptidyl peptidase IV in complex with 8-(3-aminopiperidin-1-yl)-7-[(2-bromophenyl)methyl]-1,3-dimethyl-2,3,6,7-tetrahydro-1H-purine-2,6-dione [Sus scrofa] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q4I8Q4 | 5.70e-08 | 51 | 233 | 21 | 215 | Acyl-protein thioesterase 1 OS=Gibberella zeae (strain ATCC MYA-4620 / CBS 123657 / FGSC 9075 / NRRL 31084 / PH-1) OX=229533 GN=FGRAMPH1_01T20223 PE=3 SV=1 |
| Q7XR62 | 8.42e-08 | 51 | 232 | 14 | 202 | Probable inactive carboxylesterase Os04g0669700 OS=Oryza sativa subsp. japonica OX=39947 GN=Os04g0669700 PE=3 SV=1 |
| D4AR77 | 3.65e-07 | 12 | 231 | 368 | 605 | Uncharacterized secreted protein ARB_06907 OS=Arthroderma benhamiae (strain ATCC MYA-4681 / CBS 112371) OX=663331 GN=ARB_06907 PE=1 SV=2 |
| B0XRV0 | 1.11e-06 | 36 | 250 | 458 | 697 | Dipeptidyl-peptidase 5 OS=Neosartorya fumigata (strain CEA10 / CBS 144.89 / FGSC A1163) OX=451804 GN=AFUB_024920 PE=1 SV=1 |
| B8NBM3 | 1.11e-06 | 36 | 250 | 461 | 700 | Probable dipeptidyl-peptidase 5 OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=dpp5 PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
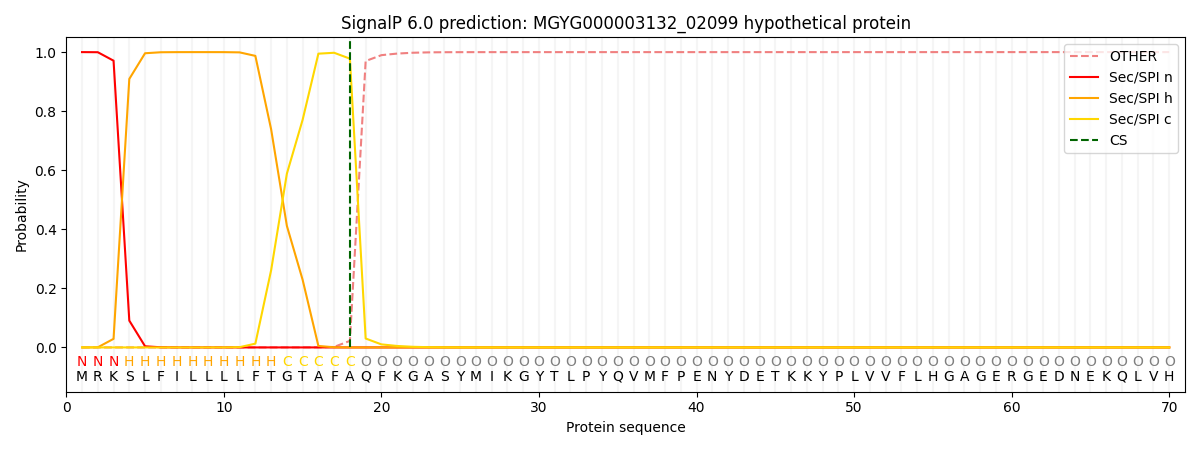
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000365 | 0.998914 | 0.000196 | 0.000159 | 0.000159 | 0.000153 |

