You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003133_01040
You are here: Home > Sequence: MGYG000003133_01040
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Niallia sp900199695 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Bacillales_B; DSM-18226; Niallia; Niallia sp900199695 | |||||||||||
| CAZyme ID | MGYG000003133_01040 | |||||||||||
| CAZy Family | GH73 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 43216; End: 47100 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH73 | 1141 | 1276 | 2.2e-24 | 0.9453125 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG4193 | LytD | 5.44e-75 | 1083 | 1294 | 48 | 245 | Beta- N-acetylglucosaminidase [Carbohydrate transport and metabolism]. |
| smart00047 | LYZ2 | 5.55e-30 | 1127 | 1284 | 1 | 146 | Lysozyme subfamily 2. Eubacterial enzymes distantly related to eukaryotic lysozymes. |
| pfam07691 | PA14 | 1.67e-11 | 150 | 274 | 7 | 129 | PA14 domain. This domain forms an insert in bacterial beta-glucosidases and is found in other glycosidases, glycosyltransferases, proteases, amidases, yeast adhesins, and bacterial toxins, including anthrax protective antigen (PA). The domain also occurs in a Dictyostelium prespore-cell-inducing factor Psi and in fibrocystin, the mammalian protein whose mutation leads to polycystic kidney and hepatic disease. The crystal structure of PA shows that this domain (named PA14 after its location in the PA20 pro-peptide) has a beta-barrel structure. The PA14 domain sequence suggests a binding function, rather than a catalytic role. The PA14 domain distribution is compatible with carbohydrate binding. |
| smart00758 | PA14 | 2.84e-11 | 150 | 274 | 5 | 125 | domain in bacterial beta-glucosidases other glycosidases, glycosyltransferases, proteases, amidases, yeast adhesins, and bacterial toxins. |
| smart00758 | PA14 | 1.96e-08 | 835 | 948 | 8 | 117 | domain in bacterial beta-glucosidases other glycosidases, glycosyltransferases, proteases, amidases, yeast adhesins, and bacterial toxins. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QJX63823.1 | 0.0 | 1 | 1294 | 1 | 1294 |
| AYV73728.1 | 0.0 | 10 | 1294 | 1 | 1285 |
| AYV67914.1 | 0.0 | 9 | 1294 | 3 | 1288 |
| AKM20437.1 | 0.0 | 8 | 1294 | 23 | 1279 |
| APH03558.1 | 2.19e-200 | 43 | 956 | 5 | 1142 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6FXO_A | 8.63e-51 | 1083 | 1294 | 37 | 244 | ChainA, Bifunctional autolysin [Staphylococcus aureus subsp. aureus Mu50] |
| 4PI7_A | 5.72e-45 | 1083 | 1294 | 33 | 228 | ChainA, Autolysin E [Staphylococcus aureus subsp. aureus Mu50],4PI9_A Chain A, Autolysin E [Staphylococcus aureus subsp. aureus Mu50],4PIA_A Chain A, Autolysin E [Staphylococcus aureus subsp. aureus Mu50] |
| 4PI8_A | 3.65e-44 | 1083 | 1294 | 33 | 228 | ChainA, Autolysin E [Staphylococcus aureus subsp. aureus Mu50] |
| 6FXP_A | 4.21e-44 | 1079 | 1282 | 42 | 235 | ChainA, Uncharacterized protein [Staphylococcus aureus subsp. aureus Mu50],6FXP_B Chain B, Uncharacterized protein [Staphylococcus aureus subsp. aureus Mu50] |
| 6U0O_B | 1.02e-43 | 1079 | 1282 | 72 | 265 | ChainB, LYZ2 domain-containing protein [Staphylococcus aureus subsp. aureus NCTC 8325] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P39848 | 2.56e-63 | 968 | 1294 | 600 | 880 | Beta-N-acetylglucosaminidase OS=Bacillus subtilis (strain 168) OX=224308 GN=lytD PE=1 SV=1 |
| Q5HQB9 | 5.40e-48 | 818 | 1294 | 839 | 1335 | Bifunctional autolysin OS=Staphylococcus epidermidis (strain ATCC 35984 / RP62A) OX=176279 GN=atl PE=3 SV=1 |
| O33635 | 5.40e-48 | 818 | 1294 | 839 | 1335 | Bifunctional autolysin OS=Staphylococcus epidermidis OX=1282 GN=atl PE=1 SV=1 |
| Q8CPQ1 | 9.36e-48 | 818 | 1294 | 839 | 1335 | Bifunctional autolysin OS=Staphylococcus epidermidis (strain ATCC 12228 / FDA PCI 1200) OX=176280 GN=atl PE=3 SV=1 |
| Q6GAG0 | 3.53e-45 | 962 | 1294 | 939 | 1250 | Bifunctional autolysin OS=Staphylococcus aureus (strain MSSA476) OX=282459 GN=atl PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
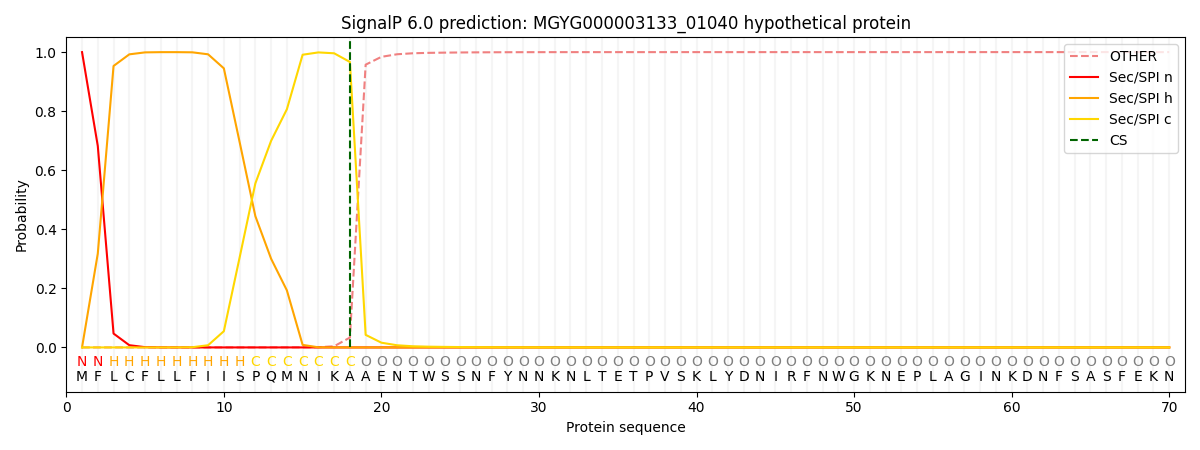
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000478 | 0.998871 | 0.000164 | 0.000166 | 0.000148 | 0.000143 |
