You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003135_01568
You are here: Home > Sequence: MGYG000003135_01568
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
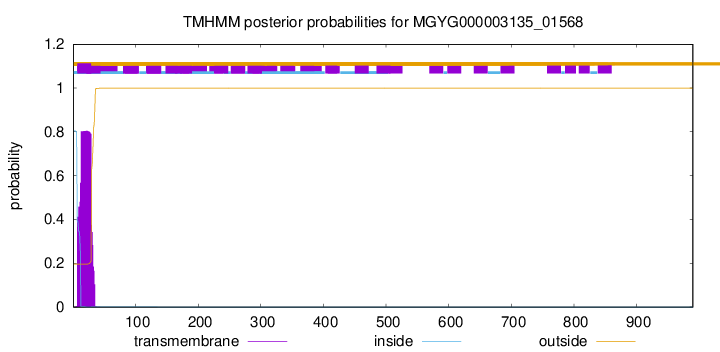
TMHMM annotations
Basic Information help
| Species | Cutibacterium granulosum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Propionibacteriales; Propionibacteriaceae; Cutibacterium; Cutibacterium granulosum | |||||||||||
| CAZyme ID | MGYG000003135_01568 | |||||||||||
| CAZy Family | CBM5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 16485; End: 19457 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| NF033681 | ExeM_NucH_DNase | 0.0 | 254 | 820 | 1 | 545 | ExeM/NucH family extracellular endonuclease. |
| COG2374 | COG2374 | 1.92e-156 | 13 | 824 | 4 | 792 | Predicted extracellular nuclease [General function prediction only]. |
| NF033680 | exonuc_ExeM-GG | 5.74e-61 | 13 | 837 | 3 | 861 | extracellular exonuclease ExeM. ExeM, as described in Shewanella oneidensis, is a biofilm formation-associated exonuclease that cleaves extracellular DNA (eDNA), a biofilm component. Members of the ExeM family contain two or three pairs of Cys residues, presumed to form disulfide bonds, and a C-terminal GlyGly-CTERM membrane-anchoring segment. Strangely, engineered removal of the GlyGly-CTERM region did not result in net export from the cell and appearance of the enzyme in culture supernatants. |
| cd10283 | MnuA_DNase1-like | 9.73e-58 | 499 | 818 | 1 | 266 | Mycoplasma pulmonis MnuA nuclease-like. This subfamily includes Mycoplasma pulmonis MnuA, a membrane-associated nuclease related to Deoxyribonuclease 1 (DNase1 or DNase I, EC 3.1.21.1). The in vivo role of MnuA is as yet undetermined. This subfamily belongs to the large EEP (exonuclease/endonuclease/phosphatase) superfamily that contains functionally diverse enzymes that share a common catalytic mechanism of cleaving phosphodiester bonds. |
| cd04486 | YhcR_OBF_like | 1.96e-16 | 253 | 330 | 1 | 73 | YhcR_OBF_like: A subfamily of OB-fold domains similar to the OB folds of Bacillus subtilis YhcR. YhcR is a sugar-nonspecific nuclease, which is active in the presence of Ca2+ and Mn2+. It cleaves RNA endonucleolytically, producing 3'-monophosphate nucleosides. YhcR appears to be the major Ca2+ activated nuclease of B. subtilis. YhcR may be localized in the cell wall. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QAY59561.1 | 6.31e-195 | 41 | 822 | 35 | 930 |
| ACZ32093.1 | 2.45e-186 | 8 | 819 | 6 | 829 |
| QAY71724.1 | 8.68e-185 | 35 | 819 | 35 | 839 |
| ANC31565.1 | 6.51e-184 | 17 | 818 | 24 | 837 |
| QAY64865.1 | 8.82e-183 | 14 | 828 | 21 | 855 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
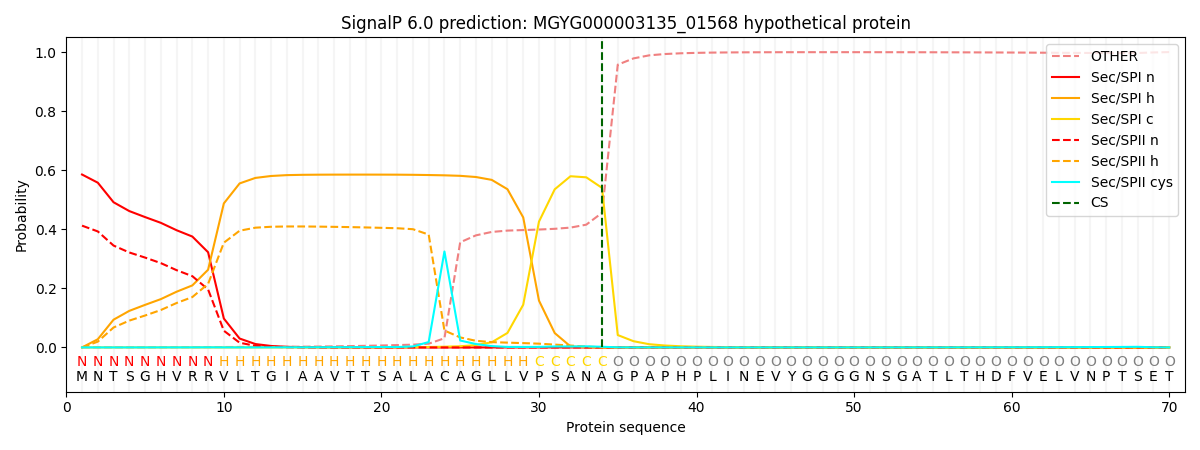
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.001233 | 0.575438 | 0.420430 | 0.002213 | 0.000425 | 0.000228 |