You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003137_03474
You are here: Home > Sequence: MGYG000003137_03474
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Bradyrhizobium sp000015165 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Proteobacteria; Alphaproteobacteria; Rhizobiales; Xanthobacteraceae; Bradyrhizobium; Bradyrhizobium sp000015165 | |||||||||||
| CAZyme ID | MGYG000003137_03474 | |||||||||||
| CAZy Family | CBM50 | |||||||||||
| CAZyme Description | Beta-barrel assembly-enhancing protease | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 29772; End: 31568 Strand: + | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| TIGR02917 | PEP_TPR_lipo | 1.93e-29 | 41 | 543 | 361 | 891 | putative PEP-CTERM system TPR-repeat lipoprotein. This protein family occurs in strictly within a subset of Gram-negative bacterial species with the proposed PEP-CTERM/exosortase system, analogous to the LPXTG/sortase system common in Gram-positive bacteria. This protein occurs in a species if and only if a transmembrane histidine kinase (TIGR02916) and a DNA-binding response regulator (TIGR02915) also occur. The present of tetratricopeptide repeats (TPR) suggests protein-protein interaction, possibly for the regulation of PEP-CTERM protein expression, since many PEP-CTERM proteins in these genomes are preceded by a proposed DNA binding site for the response regulator. |
| TIGR02917 | PEP_TPR_lipo | 1.48e-17 | 257 | 555 | 137 | 429 | putative PEP-CTERM system TPR-repeat lipoprotein. This protein family occurs in strictly within a subset of Gram-negative bacterial species with the proposed PEP-CTERM/exosortase system, analogous to the LPXTG/sortase system common in Gram-positive bacteria. This protein occurs in a species if and only if a transmembrane histidine kinase (TIGR02916) and a DNA-binding response regulator (TIGR02915) also occur. The present of tetratricopeptide repeats (TPR) suggests protein-protein interaction, possibly for the regulation of PEP-CTERM protein expression, since many PEP-CTERM proteins in these genomes are preceded by a proposed DNA binding site for the response regulator. |
| TIGR02917 | PEP_TPR_lipo | 1.64e-15 | 67 | 567 | 212 | 713 | putative PEP-CTERM system TPR-repeat lipoprotein. This protein family occurs in strictly within a subset of Gram-negative bacterial species with the proposed PEP-CTERM/exosortase system, analogous to the LPXTG/sortase system common in Gram-positive bacteria. This protein occurs in a species if and only if a transmembrane histidine kinase (TIGR02916) and a DNA-binding response regulator (TIGR02915) also occur. The present of tetratricopeptide repeats (TPR) suggests protein-protein interaction, possibly for the regulation of PEP-CTERM protein expression, since many PEP-CTERM proteins in these genomes are preceded by a proposed DNA binding site for the response regulator. |
| sd00006 | TPR | 4.82e-15 | 423 | 517 | 4 | 97 | Tetratricopeptide repeat. The Tetratricopeptide repeat (TPR) typically contains 34 amino acids and is found in a variety of organisms including bacteria, cyanobacteria, yeast, fungi, plants, and humans. It is present in a variety of proteins including those involved in chaperone, cell-cycle, transcription, and protein transport complexes. The number of TPR motifs varies among proteins. Those containing 5-6 tandem repeats generate a right-handed helical structure with an amphipathic channel that is thought to accommodate an alpha-helix of a target protein. It has been proposed that TPR proteins preferentially interact with WD-40 repeat proteins, but in many instances several TPR-proteins seem to aggregate to multi-protein complexes. |
| sd00006 | TPR | 1.84e-11 | 456 | 549 | 3 | 95 | Tetratricopeptide repeat. The Tetratricopeptide repeat (TPR) typically contains 34 amino acids and is found in a variety of organisms including bacteria, cyanobacteria, yeast, fungi, plants, and humans. It is present in a variety of proteins including those involved in chaperone, cell-cycle, transcription, and protein transport complexes. The number of TPR motifs varies among proteins. Those containing 5-6 tandem repeats generate a right-handed helical structure with an amphipathic channel that is thought to accommodate an alpha-helix of a target protein. It has been proposed that TPR proteins preferentially interact with WD-40 repeat proteins, but in many instances several TPR-proteins seem to aggregate to multi-protein complexes. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BCP52622.1 | 2.59e-113 | 47 | 506 | 49 | 508 |
| BCH26208.1 | 6.40e-113 | 6 | 506 | 7 | 507 |
| BCH34192.1 | 6.40e-113 | 6 | 506 | 7 | 507 |
| QDZ00464.1 | 1.39e-105 | 3 | 506 | 4 | 503 |
| AWC25107.1 | 3.37e-101 | 49 | 506 | 40 | 503 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7RQF_A | 4.31e-08 | 318 | 503 | 279 | 459 | ChainA, TPR repeat-containing protein PA4667 [Pseudomonas aeruginosa PAO1],7RQF_B Chain B, TPR repeat-containing protein PA4667 [Pseudomonas aeruginosa PAO1] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P42810 | 8.25e-08 | 318 | 503 | 342 | 522 | TPR repeat-containing protein PA4667 OS=Pseudomonas aeruginosa (strain ATCC 15692 / DSM 22644 / CIP 104116 / JCM 14847 / LMG 12228 / 1C / PRS 101 / PAO1) OX=208964 GN=PA4667 PE=4 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
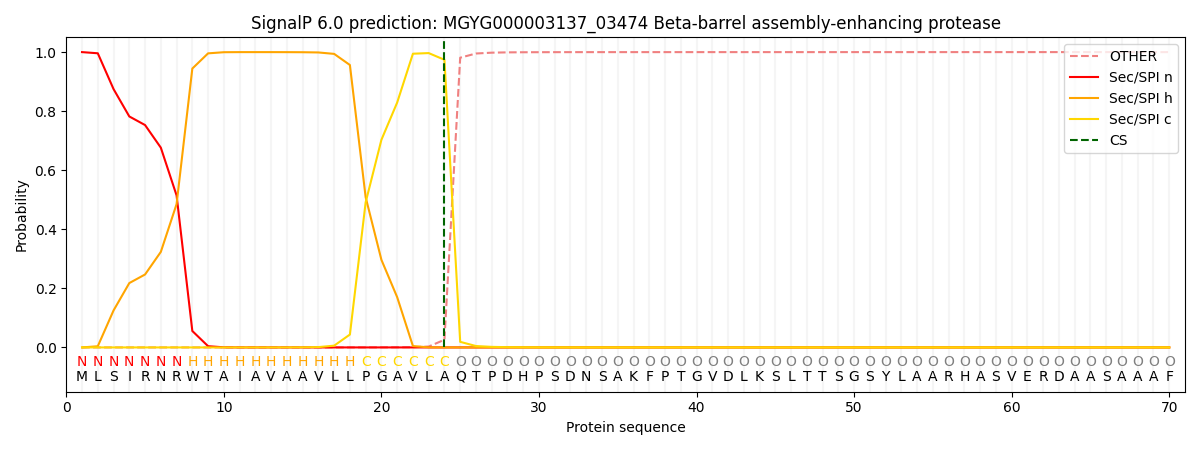
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000507 | 0.998663 | 0.000236 | 0.000205 | 0.000192 | 0.000165 |
