You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003137_06432
You are here: Home > Sequence: MGYG000003137_06432
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Bradyrhizobium sp000015165 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Proteobacteria; Alphaproteobacteria; Rhizobiales; Xanthobacteraceae; Bradyrhizobium; Bradyrhizobium sp000015165 | |||||||||||
| CAZyme ID | MGYG000003137_06432 | |||||||||||
| CAZy Family | GT41 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 22129; End: 23514 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT41 | 16 | 441 | 2.8e-117 | 0.4879432624113475 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3914 | Spy | 1.20e-117 | 82 | 441 | 257 | 620 | Predicted O-linked N-acetylglucosamine transferase, SPINDLY family [Posttranslational modification, protein turnover, chaperones]. |
| pfam13844 | Glyco_transf_41 | 5.06e-86 | 73 | 428 | 68 | 543 | Glycosyl transferase family 41. This family of glycosyltransferases includes O-linked beta-N-acetylglucosamine (O-GlcNAc) transferase, an enzyme which catalyzes the addition of O-GlcNAc to serine and threonine residues. In addition to its function as an O-GlcNAc transferase, human OGT also appears to proteolytically cleave the epigenetic cell-cycle regulator HCF-1. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ABQ39671.1 | 0.0 | 10 | 461 | 1 | 452 |
| BAM93221.1 | 9.15e-276 | 1 | 451 | 1 | 451 |
| ACK50836.1 | 5.62e-209 | 1 | 450 | 38 | 486 |
| ATQ69648.1 | 2.72e-197 | 1 | 442 | 1 | 443 |
| BBU63523.1 | 1.10e-196 | 1 | 442 | 1 | 443 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2JLB_A | 5.50e-82 | 8 | 438 | 136 | 562 | Xanthomonascampestris putative OGT (XCC0866), complex with UDP- GlcNAc phosphonate analogue [Xanthomonas campestris pv. campestris],2JLB_B Xanthomonas campestris putative OGT (XCC0866), complex with UDP- GlcNAc phosphonate analogue [Xanthomonas campestris pv. campestris],2VSY_A Xanthomonas campestris putative OGT (XCC0866), apostructure [Xanthomonas campestris pv. campestris str. ATCC 33913],2VSY_B Xanthomonas campestris putative OGT (XCC0866), apostructure [Xanthomonas campestris pv. campestris str. ATCC 33913],2XGM_A Substrate and product analogues as human O-GlcNAc transferase inhibitors. [Xanthomonas campestris],2XGM_B Substrate and product analogues as human O-GlcNAc transferase inhibitors. [Xanthomonas campestris],2XGO_A XcOGT in complex with UDP-S-GlcNAc [Xanthomonas campestris],2XGO_B XcOGT in complex with UDP-S-GlcNAc [Xanthomonas campestris],2XGS_A XcOGT in complex with C-UDP [Xanthomonas campestris],2XGS_B XcOGT in complex with C-UDP [Xanthomonas campestris] |
| 2VSN_A | 1.52e-81 | 8 | 438 | 136 | 562 | Structureand topological arrangement of an O-GlcNAc transferase homolog: insight into molecular control of intracellular glycosylation [Xanthomonas campestris pv. campestris str. 8004],2VSN_B Structure and topological arrangement of an O-GlcNAc transferase homolog: insight into molecular control of intracellular glycosylation [Xanthomonas campestris pv. campestris str. 8004] |
| 5A01_A | 4.78e-70 | 10 | 442 | 156 | 701 | O-GlcNActransferase from Drososphila melanogaster [Drosophila melanogaster],5A01_B O-GlcNAc transferase from Drososphila melanogaster [Drosophila melanogaster],5A01_C O-GlcNAc transferase from Drososphila melanogaster [Drosophila melanogaster] |
| 5NPR_A | 2.05e-62 | 78 | 447 | 224 | 709 | Thehuman O-GlcNAc transferase in complex with a thiol-linked bisubstrate inhibitor [Homo sapiens] |
| 5NPS_A | 2.08e-62 | 78 | 447 | 225 | 710 | Thehuman O-GlcNAc transferase in complex with a bisubstrate inhibitor [Homo sapiens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q9M8Y0 | 1.26e-78 | 10 | 437 | 508 | 953 | Probable UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase SEC OS=Arabidopsis thaliana OX=3702 GN=SEC PE=1 SV=1 |
| O18158 | 6.38e-68 | 76 | 442 | 654 | 1135 | UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase OS=Caenorhabditis elegans OX=6239 GN=ogt-1 PE=1 SV=2 |
| O15294 | 2.19e-60 | 78 | 447 | 548 | 1033 | UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit OS=Homo sapiens OX=9606 GN=OGT PE=1 SV=3 |
| P81436 | 2.98e-60 | 78 | 447 | 548 | 1033 | UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit OS=Oryctolagus cuniculus OX=9986 GN=OGT PE=1 SV=2 |
| Q8CGY8 | 4.07e-60 | 78 | 447 | 548 | 1033 | UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit OS=Mus musculus OX=10090 GN=Ogt PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
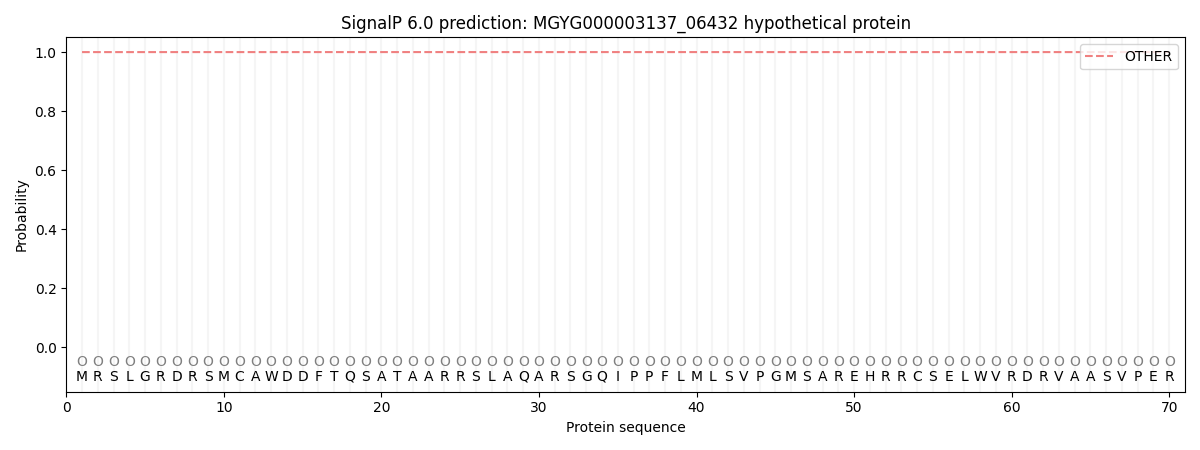
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000055 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |
