You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003137_06723
You are here: Home > Sequence: MGYG000003137_06723
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
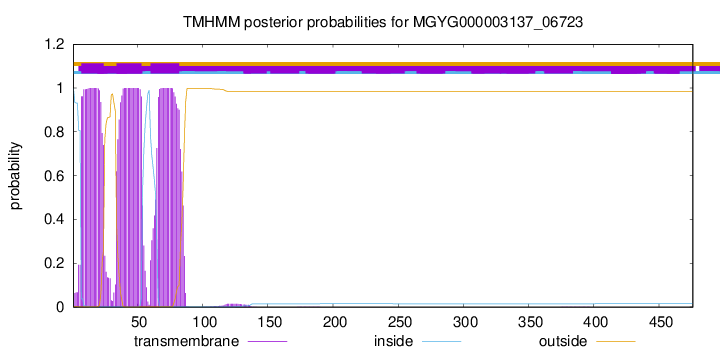
TMHMM annotations
Basic Information help
| Species | Bradyrhizobium sp000015165 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Proteobacteria; Alphaproteobacteria; Rhizobiales; Xanthobacteraceae; Bradyrhizobium; Bradyrhizobium sp000015165 | |||||||||||
| CAZyme ID | MGYG000003137_06723 | |||||||||||
| CAZy Family | GH3 | |||||||||||
| CAZyme Description | Beta-hexosaminidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 35914; End: 37344 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH3 | 156 | 389 | 1.1e-44 | 0.9722222222222222 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG1472 | BglX | 6.36e-51 | 106 | 419 | 11 | 307 | Periplasmic beta-glucosidase and related glycosidases [Carbohydrate transport and metabolism]. |
| pfam00933 | Glyco_hydro_3 | 7.49e-45 | 114 | 418 | 21 | 312 | Glycosyl hydrolase family 3 N terminal domain. |
| PRK05337 | PRK05337 | 2.25e-33 | 108 | 389 | 9 | 279 | beta-hexosaminidase; Provisional |
| PRK15098 | PRK15098 | 3.07e-04 | 213 | 471 | 147 | 391 | beta-glucosidase BglX. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ABQ36369.1 | 0.0 | 1 | 476 | 1 | 476 |
| CAL77710.1 | 1.00e-275 | 1 | 449 | 1 | 449 |
| SMX58417.1 | 2.34e-265 | 1 | 447 | 1 | 447 |
| BAM89972.1 | 5.21e-258 | 1 | 434 | 47 | 480 |
| QPF88257.1 | 7.90e-172 | 4 | 426 | 6 | 429 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6K5J_A | 5.03e-37 | 102 | 443 | 17 | 354 | Structureof a glycoside hydrolase family 3 beta-N-acetylglucosaminidase from Paenibacillus sp. str. FPU-7 [Paenibacillaceae] |
| 4YYF_A | 3.42e-32 | 91 | 423 | 33 | 354 | ChainA, Beta-N-acetylhexosaminidase [Mycolicibacterium smegmatis MC2 155],4YYF_B Chain B, Beta-N-acetylhexosaminidase [Mycolicibacterium smegmatis MC2 155],4YYF_C Chain C, Beta-N-acetylhexosaminidase [Mycolicibacterium smegmatis MC2 155] |
| 5G1M_A | 4.76e-31 | 113 | 401 | 35 | 306 | Crystalstructure of NagZ from Pseudomonas aeruginosa [Pseudomonas aeruginosa PAO1],5G1M_B Crystal structure of NagZ from Pseudomonas aeruginosa [Pseudomonas aeruginosa PAO1],5G2M_A Crystal structure of NagZ from Pseudomonas aeruginosa in complex with N-acetylglucosamine [Pseudomonas aeruginosa PAO1],5G2M_B Crystal structure of NagZ from Pseudomonas aeruginosa in complex with N-acetylglucosamine [Pseudomonas aeruginosa PAO1],5G3R_A Crystal Structure Of Nagz From Pseudomonas Aeruginosa In Complex With N-acetylglucosamine And L-ala-1,6-anhydromurnac [Pseudomonas aeruginosa PAO1],5G3R_B Crystal Structure Of Nagz From Pseudomonas Aeruginosa In Complex With N-acetylglucosamine And L-ala-1,6-anhydromurnac [Pseudomonas aeruginosa PAO1],5G5K_A Crystal structure of NagZ from Pseudomonas aeruginosa in complex with the inhibitor 2-acetamido-1,2-dideoxynojirimycin [Pseudomonas aeruginosa PAO1],5G5K_B Crystal structure of NagZ from Pseudomonas aeruginosa in complex with the inhibitor 2-acetamido-1,2-dideoxynojirimycin [Pseudomonas aeruginosa PAO1] |
| 6GFV_A | 7.06e-30 | 102 | 420 | 22 | 329 | Mtuberculosis LpqI [Mycobacterium tuberculosis H37Rv],6GFV_B M tuberculosis LpqI [Mycobacterium tuberculosis H37Rv] |
| 5G5U_A | 1.14e-29 | 113 | 401 | 35 | 306 | Crystalstructure of NagZ H174A mutant from Pseudomonas aeruginosa [Pseudomonas aeruginosa PAO1],5G5U_B Crystal structure of NagZ H174A mutant from Pseudomonas aeruginosa [Pseudomonas aeruginosa PAO1],5G6T_A Crystal structure of Zn-containing NagZ H174A mutant from Pseudomonas aeruginosa [Pseudomonas aeruginosa PAO1],5G6T_B Crystal structure of Zn-containing NagZ H174A mutant from Pseudomonas aeruginosa [Pseudomonas aeruginosa PAO1],5LY7_A Crystal structure of NagZ H174A mutant from Pseudomonas aeruginosa in complex with the inhibitor 2-acetamido-1,2-dideoxynojirimycin [Pseudomonas aeruginosa PAO1],5LY7_B Crystal structure of NagZ H174A mutant from Pseudomonas aeruginosa in complex with the inhibitor 2-acetamido-1,2-dideoxynojirimycin [Pseudomonas aeruginosa PAO1] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q9PAZ0 | 4.05e-32 | 107 | 417 | 6 | 300 | Beta-hexosaminidase OS=Xylella fastidiosa (strain 9a5c) OX=160492 GN=nagZ PE=3 SV=1 |
| Q87BR5 | 2.78e-31 | 107 | 417 | 6 | 300 | Beta-hexosaminidase OS=Xylella fastidiosa (strain Temecula1 / ATCC 700964) OX=183190 GN=nagZ PE=3 SV=1 |
| B2I6G9 | 2.78e-31 | 107 | 417 | 6 | 300 | Beta-hexosaminidase OS=Xylella fastidiosa (strain M23) OX=405441 GN=nagZ PE=3 SV=1 |
| B0U3L0 | 3.83e-31 | 107 | 417 | 6 | 300 | Beta-hexosaminidase OS=Xylella fastidiosa (strain M12) OX=405440 GN=nagZ PE=3 SV=1 |
| B7UYS5 | 1.79e-30 | 113 | 401 | 15 | 286 | Beta-hexosaminidase OS=Pseudomonas aeruginosa (strain LESB58) OX=557722 GN=nagZ PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
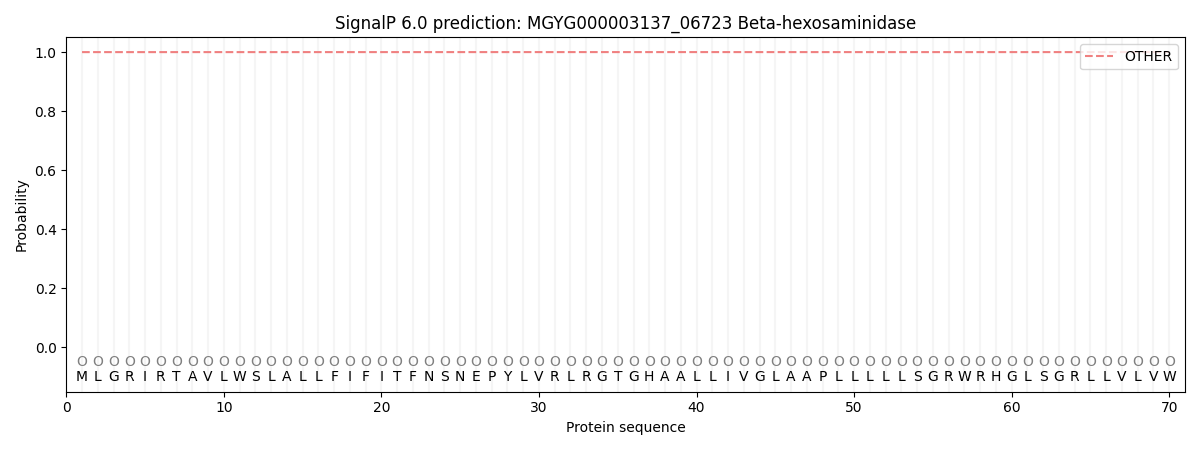
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000005 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |