You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003138_00893
You are here: Home > Sequence: MGYG000003138_00893
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Streptococcus pseudopneumoniae_O | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Lactobacillales; Streptococcaceae; Streptococcus; Streptococcus pseudopneumoniae_O | |||||||||||
| CAZyme ID | MGYG000003138_00893 | |||||||||||
| CAZy Family | GH95 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 2880; End: 8093 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH95 | 130 | 894 | 3.6e-263 | 0.9903047091412742 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam14498 | Glyco_hyd_65N_2 | 4.73e-56 | 135 | 397 | 1 | 233 | Glycosyl hydrolase family 65, N-terminal domain. This domain represents a domain found to the N-terminus of the glycosyl hydrolase 65 family catalytic domain. |
| TIGR01168 | YSIRK_signal | 7.99e-09 | 7 | 40 | 6 | 39 | Gram-positive signal peptide, YSIRK family. Many surface proteins found in Streptococcus, Staphylococcus, and related lineages share apparently homologous signal sequences. A motif resembling [YF]SIRKxxxGxxS[VIA] appears at the start of the transmembrane domain. The GxxS motif appears perfectly conserved, suggesting a specific function and not just homology. There is a strong correlation between proteins carrying this region at the N-terminus and those carrying the Gram-positive anchor domain with the LPXTG sortase processing site at the C-terminus. |
| NF033647 | adhesin_LEA | 2.79e-08 | 11 | 131 | 14 | 120 | LEA family epithelial adhesin N-terminal domain. LEA (Lactobacillus epithelium adhesin), as characterized in an adhesive commensal strain of Lactobacillus crispatus (ST1), is a large, repetitive protein with an N-terminal YSIRK-type signal peptide and a C-terminal LPXTG site for processing by sortase and attachment to the cell surface. Family members contain variable numbers of an 82 amino acid long repeats similar to Lactobacillus Rib/alpha-like repeats. This HMM describes the N-terminal region upstream of the repeat region, just over 600 amino acids long. |
| COG1554 | ATH1 | 5.56e-08 | 524 | 646 | 353 | 485 | Trehalose and maltose hydrolase (possible phosphorylase) [Carbohydrate transport and metabolism]. |
| pfam04650 | YSIRK_signal | 3.40e-07 | 8 | 31 | 1 | 24 | YSIRK type signal peptide. Many surface proteins found in Streptococcus, Staphylococcus, and related lineages share apparently homologous signal sequences. A motif resembling [YF]SIRKxxxGxxS[VIA] appears at the start of the transmembrane domain. The GxxS motif appears perfectly conserved, suggesting a specific function and not just homology. There is a strong correlation between proteins carrying this region at the N-terminus and those carrying the Gram-positive anchor domain with the LPXTG sortase processing site at the C-terminus. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QQQ35652.1 | 0.0 | 1 | 1737 | 1 | 1757 |
| AYF96278.1 | 0.0 | 1 | 1737 | 1 | 1757 |
| AQA08669.1 | 0.0 | 1 | 1737 | 1 | 1777 |
| QKL32499.1 | 0.0 | 1 | 1737 | 1 | 1817 |
| BAV79274.1 | 0.0 | 1 | 1737 | 1 | 1726 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2EAB_A | 8.02e-159 | 143 | 886 | 42 | 849 | Crystalstructure of 1,2-a-L-fucosidase from Bifidobacterium bifidum (apo form) [Bifidobacterium bifidum],2EAB_B Crystal structure of 1,2-a-L-fucosidase from Bifidobacterium bifidum (apo form) [Bifidobacterium bifidum],2EAC_A Crystal structure of 1,2-a-L-fucosidase from Bifidobacterium bifidum in complex with deoxyfuconojirimycin [Bifidobacterium bifidum],2EAC_B Crystal structure of 1,2-a-L-fucosidase from Bifidobacterium bifidum in complex with deoxyfuconojirimycin [Bifidobacterium bifidum] |
| 2EAD_A | 5.71e-158 | 143 | 886 | 42 | 849 | ChainA, Alpha-fucosidase [Bifidobacterium bifidum],2EAD_B Chain B, Alpha-fucosidase [Bifidobacterium bifidum] |
| 2EAE_A | 1.07e-157 | 143 | 886 | 41 | 848 | ChainA, Alpha-fucosidase [Bifidobacterium bifidum] |
| 2RDY_A | 2.93e-132 | 151 | 920 | 16 | 784 | ChainA, BH0842 protein [Halalkalibacterium halodurans C-125],2RDY_B Chain B, BH0842 protein [Halalkalibacterium halodurans C-125] |
| 4UFC_A | 2.78e-128 | 151 | 901 | 35 | 758 | Crystalstructure of the GH95 enzyme BACOVA_03438 [Bacteroides ovatus],4UFC_B Crystal structure of the GH95 enzyme BACOVA_03438 [Bacteroides ovatus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q8L7W8 | 8.31e-106 | 152 | 901 | 66 | 827 | Alpha-L-fucosidase 2 OS=Arabidopsis thaliana OX=3702 GN=FUC95A PE=1 SV=1 |
| Q5AU81 | 3.23e-84 | 155 | 871 | 46 | 780 | Alpha-fucosidase A OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=afcA PE=1 SV=1 |
| A2R797 | 1.91e-83 | 144 | 871 | 30 | 761 | Probable alpha-fucosidase A OS=Aspergillus niger (strain CBS 513.88 / FGSC A1513) OX=425011 GN=afcA PE=3 SV=1 |
| Q2USL3 | 2.54e-60 | 151 | 884 | 31 | 712 | Probable alpha-fucosidase A OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=afcA PE=3 SV=2 |
| P0DTR5 | 1.96e-11 | 1012 | 1273 | 825 | 1063 | A type blood alpha-D-galactosamine galactosaminidase OS=Flavonifractor plautii OX=292800 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
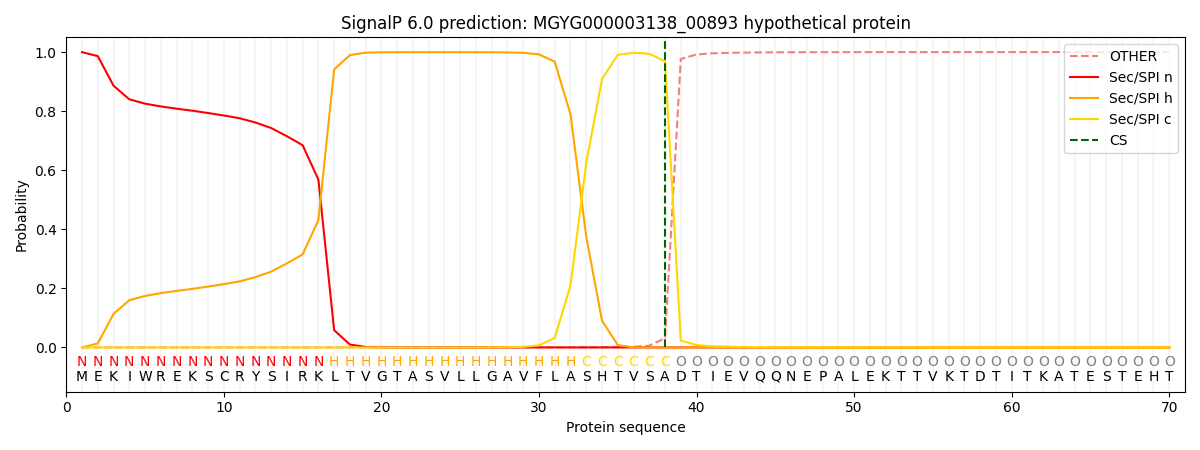
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000539 | 0.998728 | 0.000222 | 0.000180 | 0.000156 | 0.000143 |
