You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003144_01203
You are here: Home > Sequence: MGYG000003144_01203
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Aeromonas rivipollensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Proteobacteria; Gammaproteobacteria; Enterobacterales; Aeromonadaceae; Aeromonas; Aeromonas rivipollensis | |||||||||||
| CAZyme ID | MGYG000003144_01203 | |||||||||||
| CAZy Family | CE2 | |||||||||||
| CAZyme Description | Acetylxylan esterase / glucomannan deacetylase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 328882; End: 329961 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE2 | 142 | 351 | 4.2e-50 | 0.9952153110047847 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd01831 | Endoglucanase_E_like | 2.74e-30 | 142 | 352 | 1 | 169 | Endoglucanase E-like members of the SGNH hydrolase family; Endoglucanase E catalyzes the endohydrolysis of 1,4-beta-glucosidic linkages in cellulose, lichenin and cereal beta-D-glucans. |
| pfam17996 | CE2_N | 9.09e-11 | 33 | 118 | 1 | 87 | Carbohydrate esterase 2 N-terminal. This is the N-terminal beta-sheet domain with jelly roll topology found in CE2 acetyl-esterase from the bacterium Clostridium thermocellum. This enzyme displays dual activities, it catalyses the deacetylation of plant polysaccharides and also potentiates the activity of its appended cellulase catalytic module through its noncatalytic cellulose binding function. This N-terminal jelly-roll domain appears to extend the substrate/cellulose binding cleft of the catalytic domain in C.thermocellum. |
| cd00229 | SGNH_hydrolase | 3.67e-08 | 143 | 348 | 1 | 185 | SGNH_hydrolase, or GDSL_hydrolase, is a diverse family of lipases and esterases. The tertiary fold of the enzyme is substantially different from that of the alpha/beta hydrolase family and unique among all known hydrolases; its active site closely resembles the typical Ser-His-Asp(Glu) triad from other serine hydrolases, but may lack the carboxlic acid. |
| pfam13472 | Lipase_GDSL_2 | 3.28e-07 | 145 | 338 | 1 | 172 | GDSL-like Lipase/Acylhydrolase family. This family of presumed lipases and related enzymes are similar to pfam00657. |
| pfam00657 | Lipase_GDSL | 0.005 | 143 | 291 | 1 | 134 | GDSL-like Lipase/Acylhydrolase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BCS48449.1 | 7.42e-107 | 7 | 357 | 8 | 361 |
| QEO83299.1 | 1.31e-105 | 24 | 357 | 20 | 353 |
| AYO65408.1 | 1.31e-105 | 24 | 357 | 20 | 353 |
| QHU97014.1 | 1.31e-105 | 24 | 357 | 20 | 353 |
| ABO90850.1 | 1.31e-105 | 24 | 357 | 20 | 353 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2W9X_A | 9.01e-34 | 48 | 344 | 50 | 346 | Theactive site of a carbohydrate esterase displays divergent catalytic and non-catalytic binding functions [Cellvibrio japonicus],2W9X_B The active site of a carbohydrate esterase displays divergent catalytic and non-catalytic binding functions [Cellvibrio japonicus] |
| 2WAA_A | 9.04e-19 | 24 | 353 | 13 | 335 | Structureof a family two carbohydrate esterase from Cellvibrio japonicus [Cellvibrio japonicus] |
| 2WAO_A | 1.29e-15 | 50 | 354 | 29 | 330 | ChainA, ENDOGLUCANASE E [Acetivibrio thermocellus] |
| 2WAB_A | 3.19e-15 | 50 | 354 | 29 | 330 | ChainA, ENDOGLUCANASE E [Acetivibrio thermocellus] |
| 4XVH_A | 5.27e-15 | 47 | 340 | 14 | 312 | Crystalstructure of a Corynascus thermopiles (Myceliophthora fergusii) carbohydrate esterase family 2 (CE2) enzyme plus carbohydrate binding domain (CBD) [Chaetomium] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| B3PDE5 | 4.42e-33 | 48 | 344 | 50 | 346 | Acetylxylan esterase / glucomannan deacetylase OS=Cellvibrio japonicus (strain Ueda107) OX=498211 GN=ce2C PE=1 SV=1 |
| B3PIB0 | 5.52e-18 | 24 | 353 | 32 | 354 | Acetylxylan esterase / glucomannan deacetylase OS=Cellvibrio japonicus (strain Ueda107) OX=498211 GN=axe2C PE=1 SV=1 |
| P10477 | 1.96e-14 | 50 | 354 | 510 | 811 | Cellulase/esterase CelE OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=celE PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
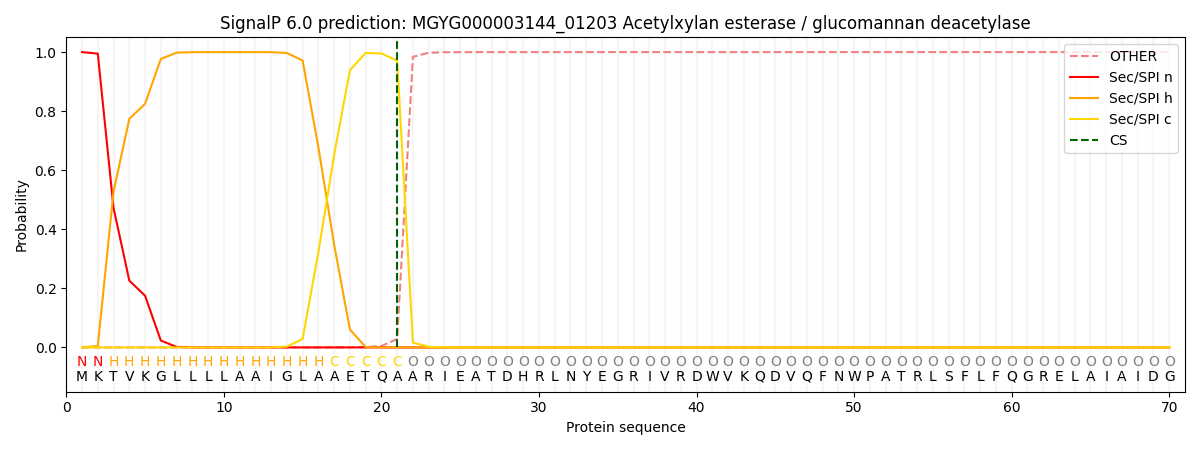
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000255 | 0.998988 | 0.000194 | 0.000196 | 0.000183 | 0.000158 |
