You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003144_03058
You are here: Home > Sequence: MGYG000003144_03058
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Aeromonas rivipollensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Proteobacteria; Gammaproteobacteria; Enterobacterales; Aeromonadaceae; Aeromonas; Aeromonas rivipollensis | |||||||||||
| CAZyme ID | MGYG000003144_03058 | |||||||||||
| CAZy Family | GH23 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 110992; End: 111576 Strand: + | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG4764 | COG4764 | 4.28e-81 | 2 | 186 | 9 | 197 | Uncharacterized protein [Function unknown]. |
| cd00442 | Lyz-like | 6.61e-08 | 51 | 122 | 1 | 59 | lysozyme-like domains. This family contains several members, including soluble lytic transglycosylases (SLT), goose egg-white lysozymes (GEWL), hen egg-white lysozymes (HEWL), chitinases, bacteriophage lambda lysozymes, endolysins, autolysins, chitosanases, and pesticin. Typical members are involved in the hydrolysis of beta-1,4- linked polysaccharides. |
| COG0741 | MltE | 5.38e-04 | 28 | 187 | 133 | 287 | Soluble lytic murein transglycosylase and related regulatory proteins (some contain LysM/invasin domains) [Cell wall/membrane/envelope biogenesis]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AHE48137.1 | 3.51e-148 | 1 | 194 | 1 | 194 |
| QIY88777.1 | 1.77e-147 | 1 | 194 | 7 | 200 |
| QYK81323.1 | 6.48e-147 | 1 | 194 | 4 | 197 |
| AVP91954.1 | 9.03e-147 | 1 | 194 | 23 | 216 |
| QJT20627.1 | 1.28e-146 | 1 | 194 | 23 | 216 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as LIPO
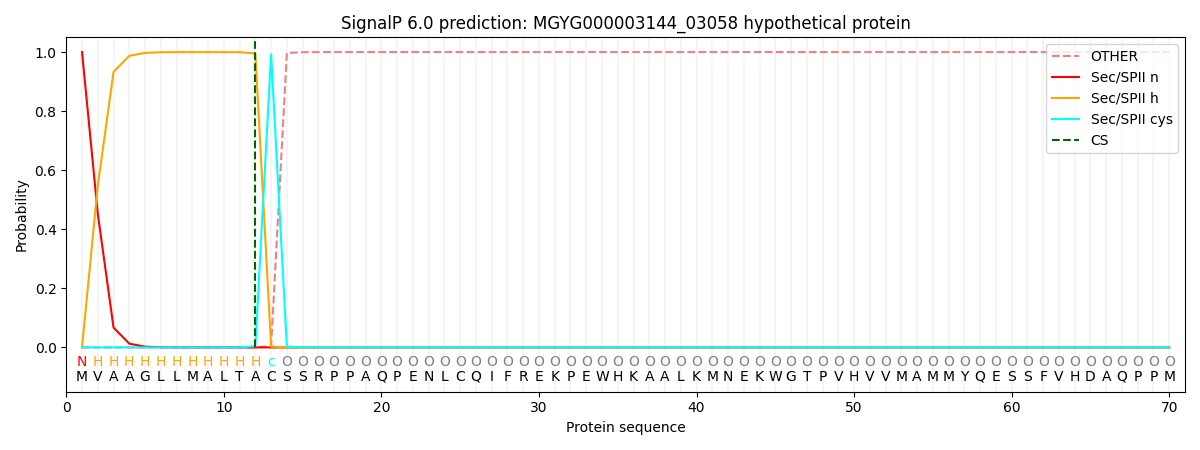
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000012 | 1.000010 | 0.000000 | 0.000000 | 0.000000 |
