You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003145_01527
You are here: Home > Sequence: MGYG000003145_01527
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
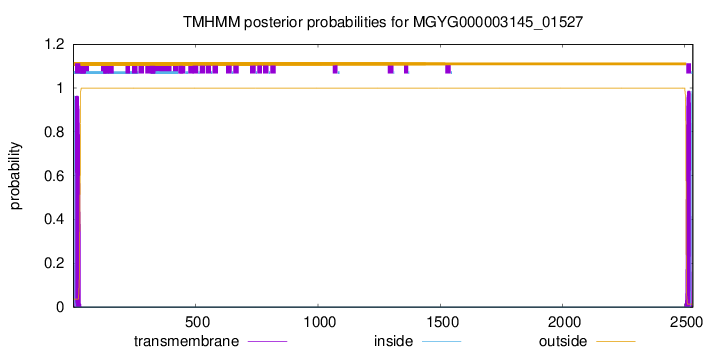
TMHMM annotations
Basic Information help
| Species | UMGS822 sp900545825 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Actinomycetales; Actinomycetaceae; UMGS822; UMGS822 sp900545825 | |||||||||||
| CAZyme ID | MGYG000003145_01527 | |||||||||||
| CAZy Family | CBM32 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 30146; End: 37747 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH35 | 1145 | 1472 | 1.8e-86 | 0.9869706840390879 |
| GH20 | 325 | 681 | 5.3e-85 | 0.973293768545994 |
| CBM32 | 54 | 178 | 9.6e-22 | 0.9274193548387096 |
| CBM32 | 963 | 1088 | 1.3e-20 | 0.8870967741935484 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd06568 | GH20_SpHex_like | 2.71e-127 | 329 | 693 | 1 | 329 | A subgroup of the Glycosyl hydrolase family 20 (GH20) catalytic domain found in proteins similar to the N-acetylhexosaminidase from Streptomyces plicatus (SpHex). SpHex catalyzes the hydrolysis of N-acetyl-beta-hexosaminides. An Asp residue within the active site plays a critical role in substrate-assisted catalysis by orienting the 2-acetamido group and stabilizing the transition state. The GH20 hexosaminidases are thought to act via a catalytic mechanism in which the catalytic nucleophile is not provided by solvent or the enzyme, but by the substrate itself. Proteins belonging to this subgroup lack the C-terminal PKD (polycystic kidney disease I)-like domain found in the chitobiases. |
| cd06563 | GH20_chitobiase-like | 2.33e-90 | 329 | 693 | 1 | 357 | The chitobiase of Serratia marcescens is a beta-N-1,4-acetylhexosaminidase with a glycosyl hydrolase family 20 (GH20) domain that hydrolyzes the beta-1,4-glycosidic linkages in oligomers derived from chitin. Chitin is degraded by a two step process: i) a chitinase hydrolyzes the chitin to oligosaccharides and disaccharides such as di-N-acetyl-D-glucosamine and chitobiose, ii) chitobiase then further degrades these oligomers into monomers. This GH20 domain family includes an N-acetylglucosamidase (GlcNAcase A) from Pseudoalteromonas piscicida and an N-acetylhexosaminidase (SpHex) from Streptomyces plicatus. SpHex lacks the C-terminal PKD (polycystic kidney disease I)-like domain found in the chitobiases. The GH20 hexosaminidases are thought to act via a catalytic mechanism in which the catalytic nucleophile is not provided by solvent or the enzyme, but by the substrate itself. |
| pfam01301 | Glyco_hydro_35 | 1.63e-83 | 1144 | 1473 | 1 | 315 | Glycosyl hydrolases family 35. |
| pfam00728 | Glyco_hydro_20 | 3.57e-82 | 329 | 680 | 1 | 345 | Glycosyl hydrolase family 20, catalytic domain. This domain has a TIM barrel fold. |
| cd02742 | GH20_hexosaminidase | 7.20e-59 | 331 | 678 | 1 | 302 | Beta-N-acetylhexosaminidases of glycosyl hydrolase family 20 (GH20) catalyze the removal of beta-1,4-linked N-acetyl-D-hexosamine residues from the non-reducing ends of N-acetyl-beta-D-hexosaminides including N-acetylglucosides and N-acetylgalactosides. These enzymes are broadly distributed in microorganisms, plants and animals, and play roles in various key physiological and pathological processes. These processes include cell structural integrity, energy storage, cellular signaling, fertilization, pathogen defense, viral penetration, the development of carcinomas, inflammatory events and lysosomal storage disorders. The GH20 enzymes include the eukaryotic beta-N-acetylhexosaminidases A and B, the bacterial chitobiases, dispersin B, and lacto-N-biosidase. The GH20 hexosaminidases are thought to act via a catalytic mechanism in which the catalytic nucleophile is not provided by the solvent or the enzyme, but by the substrate itself. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| VEG55570.1 | 0.0 | 1117 | 2371 | 58 | 1341 |
| QQM67568.1 | 0.0 | 1117 | 2371 | 57 | 1341 |
| QWW19060.1 | 0.0 | 1113 | 2531 | 31 | 1492 |
| AOZ72117.1 | 0.0 | 1122 | 2473 | 54 | 1414 |
| QQO77161.1 | 0.0 | 3 | 1100 | 9 | 1119 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4C7D_A | 2.43e-88 | 183 | 715 | 1 | 493 | Structureand activity of the GH20 beta-N-acetylhexosaminidase from Streptomyces coelicolor A3(2) [Streptomyces coelicolor],4C7D_B Structure and activity of the GH20 beta-N-acetylhexosaminidase from Streptomyces coelicolor A3(2) [Streptomyces coelicolor],4C7F_A Structure and activity of the GH20 beta-N-acetylhexosaminidase from Streptomyces coelicolor A3(2) [Streptomyces coelicolor],4C7F_B Structure and activity of the GH20 beta-N-acetylhexosaminidase from Streptomyces coelicolor A3(2) [Streptomyces coelicolor] |
| 4C7G_A | 6.10e-88 | 183 | 715 | 1 | 493 | Structureand activity of the GH20 beta-N-acetylhexosaminidase from Streptomyces coelicolor A3(2) [Streptomyces coelicolor] |
| 6YXZ_JJJ | 5.63e-87 | 28 | 770 | 6 | 772 | ChainJJJ, Beta-N-acetylhexosaminidase [Bifidobacterium bifidum] |
| 6Z14_JJJ | 1.36e-86 | 28 | 770 | 6 | 772 | ChainJJJ, Beta-N-acetylhexosaminidase [Bifidobacterium bifidum] |
| 1HP4_A | 5.44e-86 | 185 | 715 | 21 | 511 | ChainA, Beta-n-acetylhexosaminidase [Streptomyces plicatus],1HP5_A Chain A, Beta-n-acetylhexosaminidase [Streptomyces plicatus],1JAK_A Chain A, Beta-N-acetylhexosaminidase [Streptomyces plicatus],1M01_A Chain A, Beta-N-acetylhexosaminidase [Streptomyces plicatus],5FCZ_A Chain A, B-N-acetylhexosaminidase [Streptomyces plicatus],5FD0_A Chain A, B-N-acetylhexosaminidase [Streptomyces plicatus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q7WUL4 | 1.04e-72 | 190 | 722 | 1 | 492 | Beta-N-acetylhexosaminidase OS=Cellulomonas fimi OX=1708 GN=hex20 PE=1 SV=1 |
| B8N2I5 | 1.27e-64 | 1134 | 1475 | 28 | 376 | Probable beta-galactosidase C OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=lacC PE=3 SV=1 |
| Q2UMD5 | 1.27e-64 | 1134 | 1475 | 28 | 376 | Probable beta-galactosidase C OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=lacC PE=3 SV=1 |
| A1D1Z9 | 2.12e-64 | 1138 | 1546 | 46 | 453 | Probable beta-galactosidase A OS=Neosartorya fischeri (strain ATCC 1020 / DSM 3700 / CBS 544.65 / FGSC A1164 / JCM 1740 / NRRL 181 / WB 181) OX=331117 GN=lacA PE=3 SV=1 |
| Q4WS33 | 2.12e-64 | 1138 | 1546 | 46 | 453 | Probable beta-galactosidase A OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=lacA PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
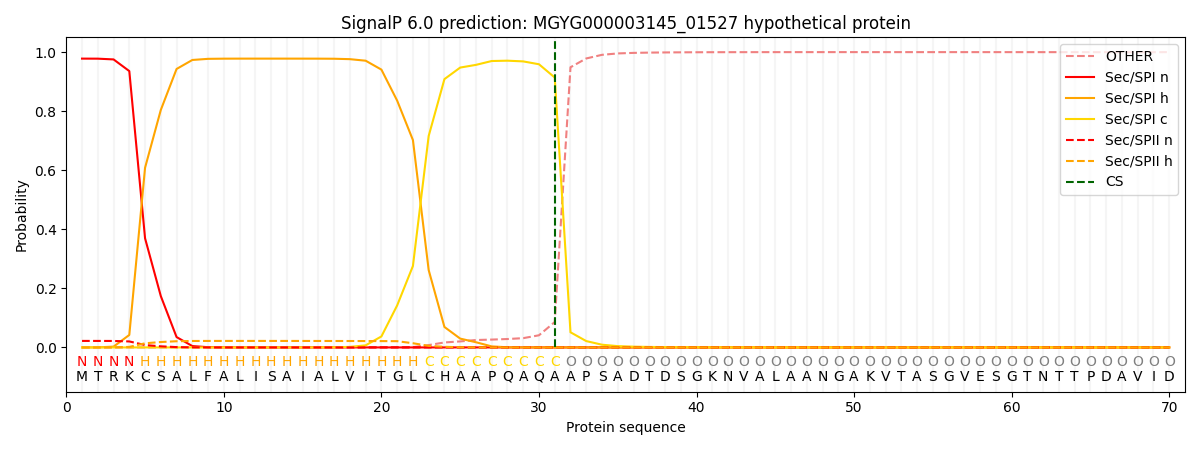
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000509 | 0.974992 | 0.023746 | 0.000304 | 0.000218 | 0.000181 |