You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003147_00345
You are here: Home > Sequence: MGYG000003147_00345
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Robinsoniella sp900555455 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Robinsoniella; Robinsoniella sp900555455 | |||||||||||
| CAZyme ID | MGYG000003147_00345 | |||||||||||
| CAZy Family | GH36 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 434998; End: 438849 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH36 | 275 | 529 | 3.3e-31 | 0.35755813953488375 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd14791 | GH36 | 1.36e-33 | 305 | 554 | 6 | 244 | glycosyl hydrolase family 36 (GH36). GH36 enzymes occur in prokaryotes, eukaryotes, and archaea with a wide range of hydrolytic activities, including alpha-galactosidase, alpha-N-acetylgalactosaminidase, stachyose synthase, and raffinose synthase. All GH36 enzymes cleave a terminal carbohydrate moiety from a substrate that varies considerably in size, depending on the enzyme, and may be either a starch or a glycoprotein. GH36 members are retaining enzymes that cleave their substrates via an acid/base-catalyzed, double-displacement mechanism involving a covalent glycosyl-enzyme intermediate. Two aspartic acid residues have been identified as the catalytic nucleophile and the acid/base, respectively. |
| pfam02065 | Melibiase | 1.67e-16 | 310 | 528 | 48 | 262 | Melibiase. Glycoside hydrolase families GH27, GH31 and GH36 form the glycoside hydrolase clan GH-D. Glycoside hydrolase family 36 can be split into 11 families, GH36A to GH36K. This family includes enzymes from GH36A-B and GH36D-K and from GH27. |
| COG3345 | GalA | 2.96e-14 | 288 | 401 | 274 | 396 | Alpha-galactosidase [Carbohydrate transport and metabolism]. |
| cd00063 | FN3 | 3.95e-10 | 1108 | 1187 | 1 | 89 | Fibronectin type 3 domain; One of three types of internal repeats found in the plasma protein fibronectin. Its tenth fibronectin type III repeat contains an RGD cell recognition sequence in a flexible loop between 2 strands. Approximately 2% of all animal proteins contain the FN3 repeat; including extracellular and intracellular proteins, membrane spanning cytokine receptors, growth hormone receptors, tyrosine phosphatase receptors, and adhesion molecules. FN3-like domains are also found in bacterial glycosyl hydrolases. |
| smart00060 | FN3 | 3.85e-08 | 1108 | 1181 | 1 | 83 | Fibronectin type 3 domain. One of three types of internal repeat within the plasma protein, fibronectin. The tenth fibronectin type III repeat contains a RGD cell recognition sequence in a flexible loop between 2 strands. Type III modules are present in both extracellular and intracellular proteins. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QIA30399.1 | 0.0 | 36 | 821 | 46 | 824 |
| QQR05374.1 | 0.0 | 36 | 821 | 46 | 824 |
| ANU41754.1 | 0.0 | 36 | 821 | 46 | 824 |
| ABF39669.1 | 1.12e-83 | 28 | 687 | 32 | 681 |
| ARU29327.1 | 6.16e-83 | 77 | 690 | 76 | 689 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6PHU_A | 6.20e-10 | 283 | 401 | 330 | 447 | SpAgawild type apo structure [Streptococcus pneumoniae TIGR4],6PHV_A Chain A, Alpha-galactosidase [Streptococcus pneumoniae TIGR4] |
| 6PHW_A | 6.20e-10 | 283 | 401 | 330 | 447 | ChainA, Alpha-galactosidase [Streptococcus pneumoniae TIGR4],6PHX_A SpAga D472N structure in complex with raffinose [Streptococcus pneumoniae TIGR4],6PHY_A Chain A, Alpha-galactosidase [Streptococcus pneumoniae TIGR4],6PI0_A AgaD472N-Linear Blood group B type 2 trisaccharide complex structure [Streptococcus pneumoniae TIGR4] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P0DTR5 | 0.0 | 36 | 821 | 46 | 824 | A type blood alpha-D-galactosamine galactosaminidase OS=Flavonifractor plautii OX=292800 PE=1 SV=1 |
| P0DTR4 | 3.70e-19 | 699 | 821 | 647 | 769 | A type blood N-acetyl-alpha-D-galactosamine deacetylase OS=Flavonifractor plautii OX=292800 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
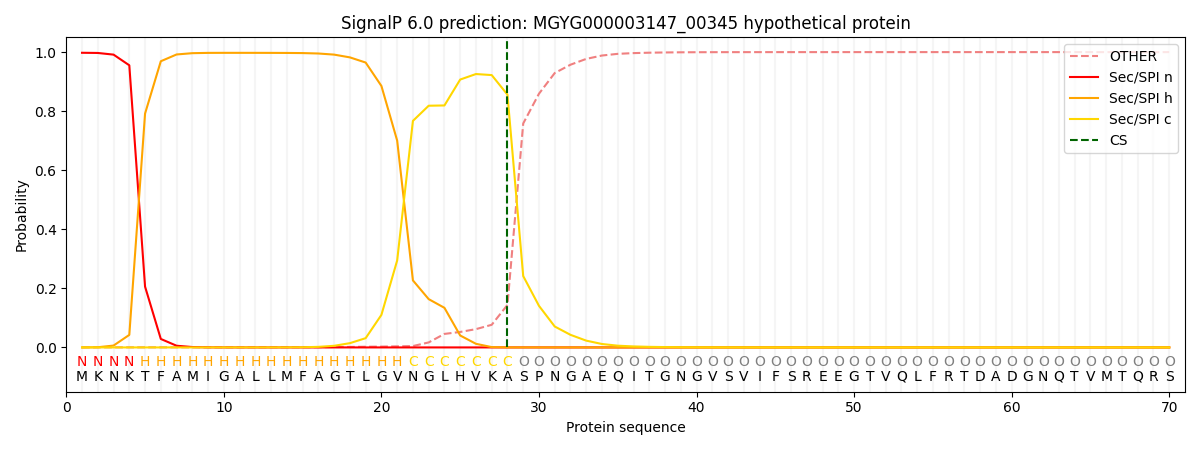
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.001259 | 0.995546 | 0.002508 | 0.000240 | 0.000206 | 0.000195 |
