You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003147_03324
You are here: Home > Sequence: MGYG000003147_03324
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
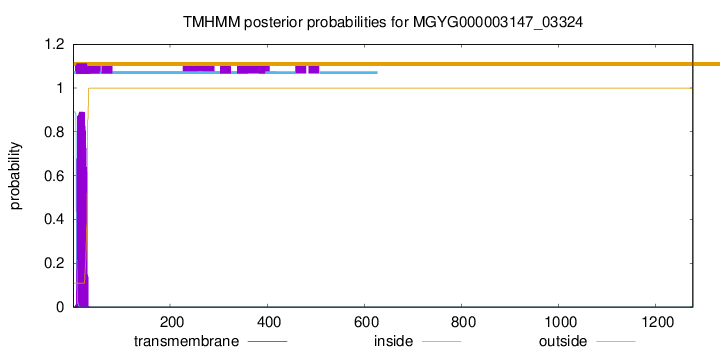
TMHMM annotations
Basic Information help
| Species | Robinsoniella sp900555455 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Robinsoniella; Robinsoniella sp900555455 | |||||||||||
| CAZyme ID | MGYG000003147_03324 | |||||||||||
| CAZy Family | GH136 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 42270; End: 46103 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH136 | 36 | 553 | 8.5e-112 | 0.9918533604887984 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG5492 | YjdB | 9.75e-16 | 868 | 1085 | 123 | 329 | Uncharacterized conserved protein YjdB, contains Ig-like domain [General function prediction only]. |
| COG5492 | YjdB | 1.11e-13 | 1004 | 1119 | 175 | 300 | Uncharacterized conserved protein YjdB, contains Ig-like domain [General function prediction only]. |
| pfam02368 | Big_2 | 2.26e-13 | 1011 | 1088 | 1 | 77 | Bacterial Ig-like domain (group 2). This family consists of bacterial domains with an Ig-like fold. Members of this family are found in bacterial and phage surface proteins such as intimins. |
| pfam02368 | Big_2 | 1.47e-11 | 836 | 913 | 1 | 77 | Bacterial Ig-like domain (group 2). This family consists of bacterial domains with an Ig-like fold. Members of this family are found in bacterial and phage surface proteins such as intimins. |
| pfam02368 | Big_2 | 4.40e-11 | 927 | 1002 | 3 | 77 | Bacterial Ig-like domain (group 2). This family consists of bacterial domains with an Ig-like fold. Members of this family are found in bacterial and phage surface proteins such as intimins. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QMW76217.1 | 0.0 | 24 | 831 | 22 | 827 |
| QIB55915.1 | 0.0 | 24 | 831 | 22 | 827 |
| QRO38181.1 | 0.0 | 1 | 831 | 1 | 831 |
| QBF74993.1 | 0.0 | 1 | 831 | 1 | 831 |
| ANY71259.1 | 7.25e-87 | 16 | 927 | 7 | 918 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7V6M_A | 2.98e-56 | 29 | 558 | 1 | 579 | ChainA, Fibronectin type III domain-containing protein [Tyzzerella nexilis] |
| 7V6I_A | 3.46e-31 | 32 | 559 | 9 | 612 | ChainA, Lacto-N-biosidase [Bifidobacterium saguini DSM 23967] |
| 6KQT_A | 6.09e-30 | 28 | 390 | 233 | 617 | CrystalStructure of GH136 lacto-N-biosidase from Eubacterium ramulus - native protein [Eubacterium ramulus ATCC 29099] |
| 6KQS_A | 5.83e-29 | 28 | 390 | 233 | 617 | CrystalStructure of GH136 lacto-N-biosidase from Eubacterium ramulus - selenomethionine derivative [Eubacterium ramulus ATCC 29099] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P33747 | 4.13e-16 | 803 | 1004 | 5 | 201 | Uncharacterized protein CA_P0160 OS=Clostridium acetobutylicum (strain ATCC 824 / DSM 792 / JCM 1419 / LMG 5710 / VKM B-1787) OX=272562 GN=CA_P0160 PE=3 SV=2 |
| P85991 | 1.15e-14 | 821 | 1005 | 79 | 258 | Ig-like virion protein OS=Serratia phage KSP90 OX=552528 PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
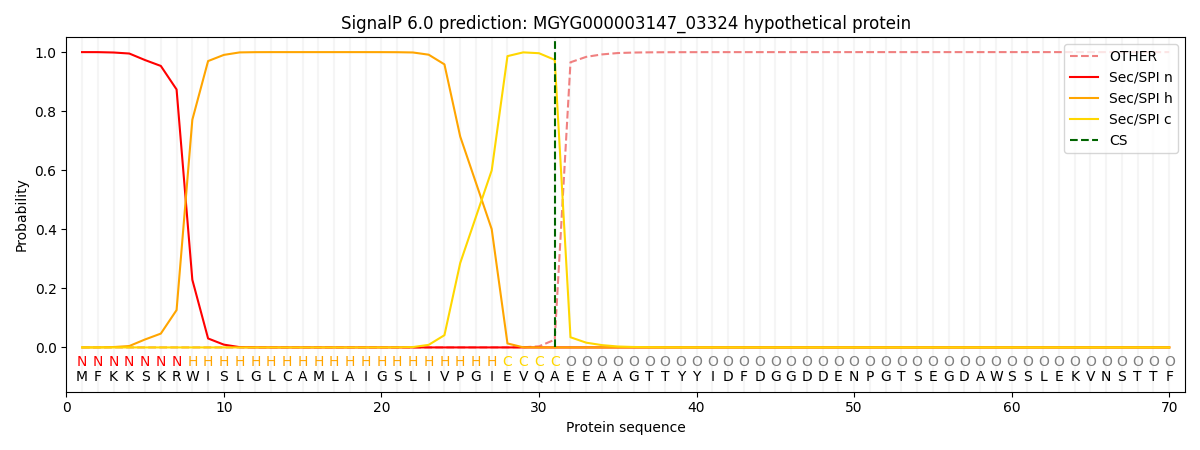
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000368 | 0.998896 | 0.000189 | 0.000199 | 0.000169 | 0.000151 |