You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003148_00112
You are here: Home > Sequence: MGYG000003148_00112
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
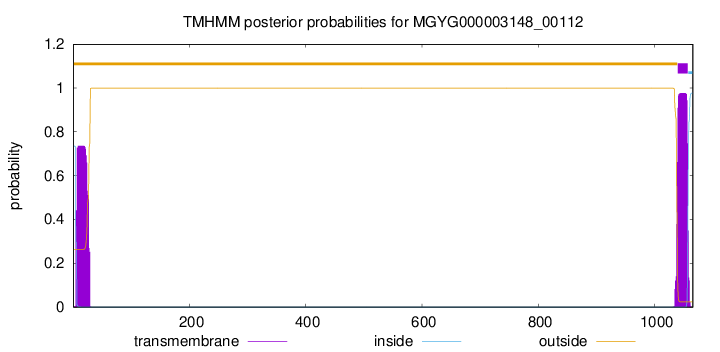
TMHMM annotations
Basic Information help
| Species | Faecalimonas sp900551895 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Faecalimonas; Faecalimonas sp900551895 | |||||||||||
| CAZyme ID | MGYG000003148_00112 | |||||||||||
| CAZy Family | GH84 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 122892; End: 126092 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH84 | 187 | 484 | 6.2e-109 | 0.9932203389830508 |
| CBM32 | 724 | 849 | 1.7e-22 | 0.9435483870967742 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam07555 | NAGidase | 1.80e-147 | 187 | 483 | 1 | 293 | beta-N-acetylglucosaminidase. This family has previously been described as a hyaluronidase. However, more recently it has been shown that this family has beta-N-acetylglucosaminidase activity. |
| pfam02838 | Glyco_hydro_20b | 1.84e-19 | 38 | 180 | 3 | 123 | Glycosyl hydrolase family 20, domain 2. This domain has a zincin-like fold. |
| pfam00754 | F5_F8_type_C | 4.26e-17 | 725 | 845 | 1 | 124 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| COG3291 | COG3291 | 8.52e-16 | 627 | 842 | 80 | 297 | PKD repeat [Function unknown]. |
| pfam00801 | PKD | 1.87e-07 | 632 | 699 | 1 | 69 | PKD domain. This domain was first identified in the Polycystic kidney disease protein PKD1. This domain has been predicted to contain an Ig-like fold. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QCJ44459.1 | 1.90e-312 | 1 | 938 | 1 | 933 |
| AZU64318.1 | 8.98e-311 | 1 | 942 | 1 | 938 |
| AQW23813.1 | 1.67e-192 | 1 | 643 | 1 | 641 |
| QUD74481.1 | 4.66e-192 | 1 | 643 | 1 | 641 |
| AXH52449.1 | 4.66e-192 | 1 | 643 | 1 | 641 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2CBI_A | 4.10e-197 | 38 | 626 | 15 | 591 | Structureof the Clostridium perfringens NagJ family 84 glycoside hydrolase, a homologue of human O-GlcNAcase [Clostridium perfringens],2CBI_B Structure of the Clostridium perfringens NagJ family 84 glycoside hydrolase, a homologue of human O-GlcNAcase [Clostridium perfringens],2CBJ_A Structure of the Clostridium perfringens NagJ family 84 glycoside hydrolase, a homologue of human O-GlcNAcase in complex with PUGNAc [Clostridium perfringens],2CBJ_B Structure of the Clostridium perfringens NagJ family 84 glycoside hydrolase, a homologue of human O-GlcNAcase in complex with PUGNAc [Clostridium perfringens],2V5C_A Family 84 glycoside hydrolase from Clostridium perfringens, 2.1 Angstrom structure [Clostridium perfringens],2V5C_B Family 84 glycoside hydrolase from Clostridium perfringens, 2.1 Angstrom structure [Clostridium perfringens],2VUR_A Chemical dissection of the link between Streptozotocin, O-GlcNAc and pancreatic cell death [Clostridium perfringens],2VUR_B Chemical dissection of the link between Streptozotocin, O-GlcNAc and pancreatic cell death [Clostridium perfringens],2X0Y_A Screening-based discovery of drug-like O-GlcNAcase inhibitor scaffolds [Clostridium perfringens],2X0Y_B Screening-based discovery of drug-like O-GlcNAcase inhibitor scaffolds [Clostridium perfringens] |
| 2J62_A | 5.78e-197 | 38 | 626 | 15 | 591 | Structureof a bacterial O-glcnacase in complex with glcnacstatin [Clostridium perfringens],2J62_B Structure of a bacterial O-glcnacase in complex with glcnacstatin [Clostridium perfringens],2WB5_A GlcNAcstatins are nanomolar inhibitors of human O-GlcNAcase inducing cellular hyper-O-GlcNAcylation [Clostridium perfringens],2WB5_B GlcNAcstatins are nanomolar inhibitors of human O-GlcNAcase inducing cellular hyper-O-GlcNAcylation [Clostridium perfringens] |
| 5OXD_A | 2.00e-196 | 38 | 622 | 17 | 589 | Complexof a C. perfringens O-GlcNAcase with a fragment hit [Clostridium perfringens] |
| 6RHE_A | 3.02e-196 | 38 | 625 | 17 | 592 | CpOGAD298N in complex with hOGA-derived S-GlcNAc peptide [Clostridium perfringens ATCC 13124] |
| 2XPK_A | 3.24e-196 | 38 | 626 | 15 | 591 | Cell-penetrant,nanomolar O-GlcNAcase inhibitors selective against lysosomal hexosaminidases [Clostridium perfringens],2XPK_B Cell-penetrant, nanomolar O-GlcNAcase inhibitors selective against lysosomal hexosaminidases [Clostridium perfringens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q0TR53 | 3.65e-192 | 1 | 643 | 1 | 641 | O-GlcNAcase NagJ OS=Clostridium perfringens (strain ATCC 13124 / DSM 756 / JCM 1290 / NCIMB 6125 / NCTC 8237 / Type A) OX=195103 GN=nagJ PE=1 SV=1 |
| Q8XL08 | 7.23e-192 | 1 | 643 | 1 | 641 | O-GlcNAcase NagJ OS=Clostridium perfringens (strain 13 / Type A) OX=195102 GN=nagJ PE=1 SV=1 |
| Q89ZI2 | 5.16e-89 | 29 | 543 | 16 | 503 | O-GlcNAcase BT_4395 OS=Bacteroides thetaiotaomicron (strain ATCC 29148 / DSM 2079 / JCM 5827 / CCUG 10774 / NCTC 10582 / VPI-5482 / E50) OX=226186 GN=BT_4395 PE=1 SV=1 |
| P26831 | 7.75e-42 | 3 | 534 | 6 | 534 | Hyaluronoglucosaminidase OS=Clostridium perfringens (strain 13 / Type A) OX=195102 GN=nagH PE=1 SV=2 |
| Q8VIJ5 | 2.17e-34 | 188 | 452 | 63 | 331 | Protein O-GlcNAcase OS=Rattus norvegicus OX=10116 GN=Oga PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
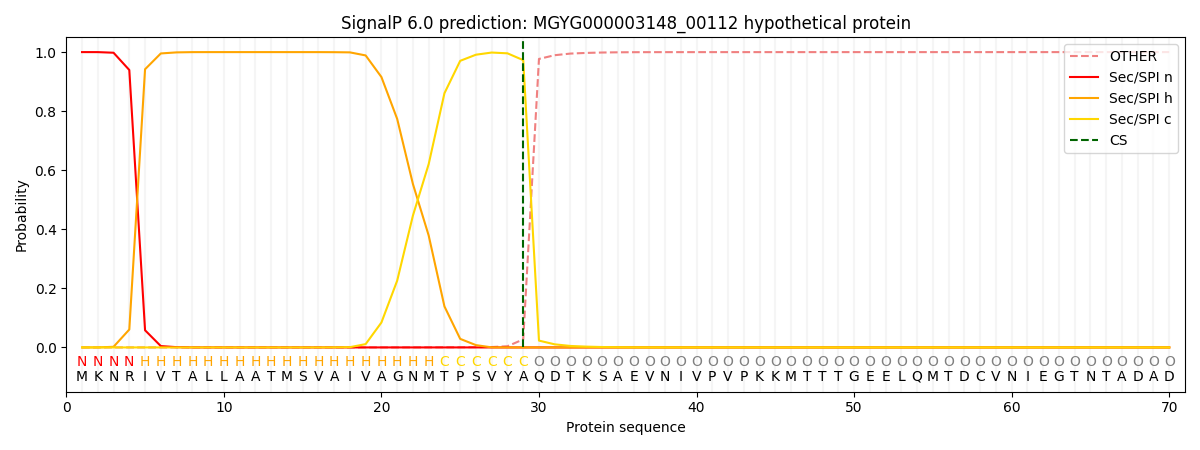
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000240 | 0.999096 | 0.000171 | 0.000169 | 0.000151 | 0.000134 |