You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003152_01934
You are here: Home > Sequence: MGYG000003152_01934
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Prevotella denticola | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotella; Prevotella denticola | |||||||||||
| CAZyme ID | MGYG000003152_01934 | |||||||||||
| CAZy Family | CBM32 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 5124; End: 5894 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM32 | 130 | 249 | 5.1e-18 | 0.8790322580645161 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00754 | F5_F8_type_C | 1.10e-08 | 139 | 249 | 17 | 127 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| cd00057 | FA58C | 5.66e-05 | 148 | 253 | 32 | 143 | Substituted updates: Jan 31, 2002 |
| smart00231 | FA58C | 4.13e-04 | 153 | 253 | 36 | 138 | Coagulation factor 5/8 C-terminal domain, discoidin domain. Cell surface-attached carbohydrate-binding domain, present in eukaryotes and assumed to have horizontally transferred to eubacterial genomes. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUB90012.1 | 2.32e-186 | 1 | 256 | 1 | 256 |
| AXV50450.1 | 6.65e-186 | 1 | 256 | 1 | 256 |
| QUB89313.1 | 9.45e-186 | 1 | 256 | 1 | 256 |
| QUB94138.1 | 9.45e-186 | 1 | 256 | 1 | 256 |
| QUI92885.1 | 9.45e-186 | 1 | 256 | 1 | 256 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
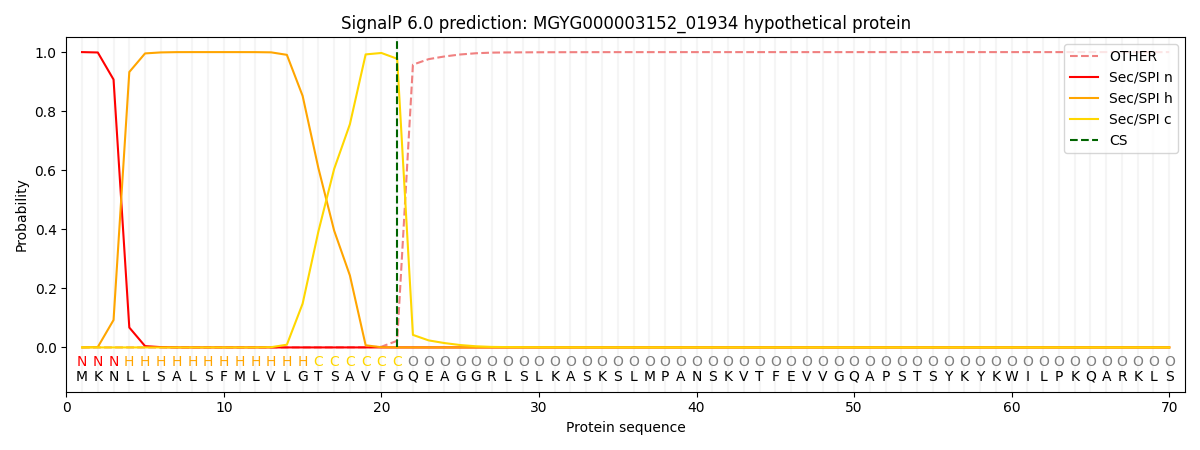
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000336 | 0.998985 | 0.000166 | 0.000175 | 0.000164 | 0.000146 |
