You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003154_01657
You are here: Home > Sequence: MGYG000003154_01657
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | UMGS1490 sp900548185 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Verrucomicrobiota; Verrucomicrobiae; Opitutales; CAG-312; UMGS1490; UMGS1490 sp900548185 | |||||||||||
| CAZyme ID | MGYG000003154_01657 | |||||||||||
| CAZy Family | GH2 | |||||||||||
| CAZyme Description | Beta-galactosidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 11778; End: 15464 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH2 | 233 | 975 | 1e-88 | 0.726063829787234 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3250 | LacZ | 6.49e-35 | 319 | 990 | 55 | 667 | Beta-galactosidase/beta-glucuronidase [Carbohydrate transport and metabolism]. |
| PRK10150 | PRK10150 | 4.00e-29 | 329 | 755 | 67 | 476 | beta-D-glucuronidase; Provisional |
| PRK10340 | ebgA | 2.35e-22 | 331 | 713 | 113 | 470 | cryptic beta-D-galactosidase subunit alpha; Reviewed |
| pfam00703 | Glyco_hydro_2 | 1.17e-16 | 448 | 555 | 1 | 106 | Glycosyl hydrolases family 2. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities. |
| pfam02836 | Glyco_hydro_2_C | 3.96e-12 | 557 | 782 | 1 | 209 | Glycosyl hydrolases family 2, TIM barrel domain. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| SDT89154.1 | 0.0 | 27 | 1224 | 23 | 1189 |
| AQQ70440.1 | 2.23e-272 | 27 | 1220 | 50 | 1231 |
| QIU96993.1 | 9.49e-266 | 51 | 1227 | 62 | 1232 |
| SDS47501.1 | 2.89e-263 | 39 | 1223 | 47 | 1203 |
| QDO68995.1 | 8.37e-263 | 23 | 1223 | 20 | 1227 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6B6L_A | 1.27e-26 | 234 | 717 | 2 | 436 | Thecrystal structure of glycosyl hydrolase family 2 (GH2) member from Bacteroides cellulosilyticus DSM 14838 [Bacteroides cellulosilyticus DSM 14838],6B6L_B The crystal structure of glycosyl hydrolase family 2 (GH2) member from Bacteroides cellulosilyticus DSM 14838 [Bacteroides cellulosilyticus DSM 14838] |
| 7RSK_A | 2.23e-26 | 234 | 717 | 2 | 436 | ChainA, Glycosyl hydrolase family 2, sugar binding domain protein [Bacteroides cellulosilyticus DSM 14838],7RSK_B Chain B, Glycosyl hydrolase family 2, sugar binding domain protein [Bacteroides cellulosilyticus DSM 14838] |
| 6XXW_A | 3.99e-24 | 324 | 758 | 69 | 476 | Structureof beta-D-Glucuronidase for Dictyoglomus thermophilum. [Dictyoglomus thermophilum H-6-12] |
| 3GM8_A | 4.41e-24 | 266 | 738 | 30 | 449 | ChainA, Glycoside hydrolase family 2, candidate beta-glycosidase [Phocaeicola vulgatus ATCC 8482] |
| 6D8K_A | 6.62e-23 | 328 | 782 | 99 | 502 | Bacteroidesmultiple species beta-glucuronidase [Bacteroides ovatus],6D8K_B Bacteroides multiple species beta-glucuronidase [Bacteroides ovatus],6D8K_C Bacteroides multiple species beta-glucuronidase [Bacteroides ovatus],6D8K_D Bacteroides multiple species beta-glucuronidase [Bacteroides ovatus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| T2KPJ7 | 1.13e-26 | 237 | 768 | 51 | 517 | Putative beta-glucuronidase OS=Formosa agariphila (strain DSM 15362 / KCTC 12365 / LMG 23005 / KMM 3901 / M-2Alg 35-1) OX=1347342 GN=BN863_21970 PE=2 SV=1 |
| T2KM09 | 1.67e-22 | 235 | 720 | 44 | 459 | Putative beta-glucuronidase OS=Formosa agariphila (strain DSM 15362 / KCTC 12365 / LMG 23005 / KMM 3901 / M-2Alg 35-1) OX=1347342 GN=BN863_22050 PE=2 SV=2 |
| P77989 | 2.33e-21 | 328 | 675 | 57 | 384 | Beta-galactosidase OS=Thermoanaerobacter pseudethanolicus (strain ATCC 33223 / 39E) OX=340099 GN=lacZ PE=3 SV=2 |
| P24131 | 2.83e-16 | 241 | 678 | 48 | 459 | Beta-galactosidase OS=Clostridium acetobutylicum OX=1488 GN=cbgA PE=2 SV=2 |
| A7LXS9 | 6.17e-16 | 231 | 717 | 36 | 499 | Beta-galactosidase BoGH2A OS=Bacteroides ovatus (strain ATCC 8483 / DSM 1896 / JCM 5824 / BCRC 10623 / CCUG 4943 / NCTC 11153) OX=411476 GN=BACOVA_02645 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
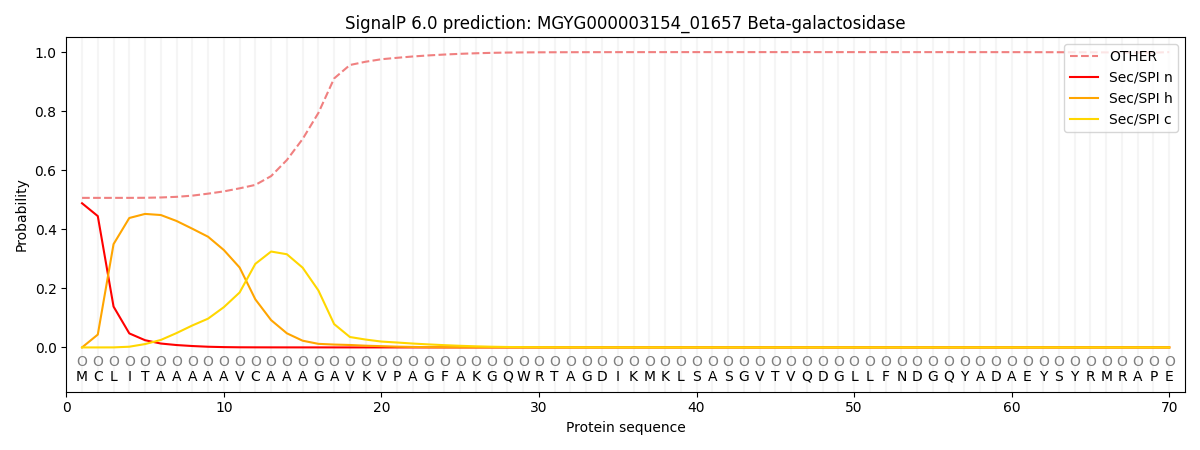
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.518701 | 0.473149 | 0.007202 | 0.000315 | 0.000266 | 0.000363 |
