You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003164_00751
You are here: Home > Sequence: MGYG000003164_00751
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
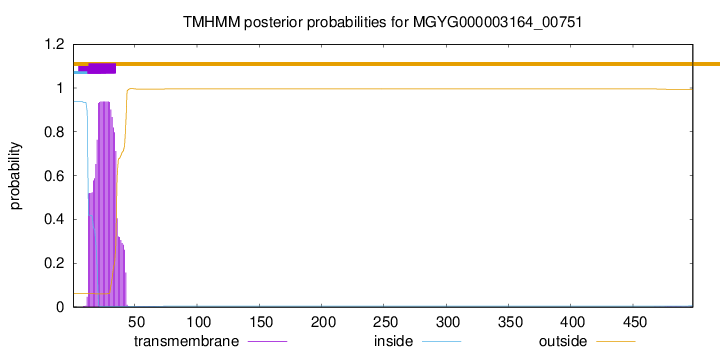
TMHMM annotations
Basic Information help
| Species | CAG-485 sp002361235 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Muribaculaceae; CAG-485; CAG-485 sp002361235 | |||||||||||
| CAZyme ID | MGYG000003164_00751 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 6600; End: 8096 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 168 | 461 | 4.9e-94 | 0.9891304347826086 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00150 | Cellulase | 6.16e-51 | 168 | 461 | 16 | 269 | Cellulase (glycosyl hydrolase family 5). |
| TIGR04056 | OMP_RagA_SusC | 7.96e-22 | 36 | 106 | 1 | 71 | TonB-linked outer membrane protein, SusC/RagA family. This model describes a distinctive clade among the TonB-linked outer membrane proteins (OMP). Members of this family are restricted to the Bacteriodetes lineage (except for Gemmatimonas aurantiaca T-27 from the novel phylum Gemmatimonadetes) and occur in high copy numbers, with over 100 members from Bacteroides thetaiotaomicron VPI-5482 alone. Published descriptions of members of this family are available for RagA from Porphyromonas gingivalis, SusC from Bacteroides thetaiotaomicron, and OmpW from Bacteroides caccae. Members form pairs with members of the SusD/RagB family (pfam07980). Transporter complexes including these outer membrane proteins are likely to import large degradation products of proteins (e.g. RagA) or carbohydrates (e.g. SusC) as nutrients, rather than siderophores. [Transport and binding proteins, Unknown substrate] |
| pfam13715 | CarbopepD_reg_2 | 6.72e-19 | 38 | 108 | 1 | 76 | CarboxypepD_reg-like domain. This domain family is found in bacteria, archaea and eukaryotes, and is approximately 90 amino acids in length. The family is found in association with pfam07715 and pfam00593. |
| COG2730 | BglC | 9.70e-19 | 155 | 466 | 54 | 365 | Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
| pfam13620 | CarboxypepD_reg | 9.13e-10 | 38 | 120 | 2 | 81 | Carboxypeptidase regulatory-like domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QRK07516.1 | 2.24e-100 | 145 | 491 | 34 | 371 |
| QDH68754.1 | 1.36e-99 | 144 | 495 | 90 | 427 |
| QIK55671.1 | 6.72e-98 | 133 | 489 | 127 | 487 |
| ALN90813.1 | 7.03e-98 | 144 | 495 | 50 | 387 |
| QIK61079.1 | 1.88e-97 | 98 | 489 | 93 | 487 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5OYC_A | 7.47e-81 | 133 | 491 | 33 | 393 | GH5endo-xyloglucanase from Cellvibrio japonicus [Cellvibrio japonicus Ueda107],5OYC_B GH5 endo-xyloglucanase from Cellvibrio japonicus [Cellvibrio japonicus Ueda107],5OYD_A GH5 endo-xyloglucanase from Cellvibrio japonicus [Cellvibrio japonicus Ueda107],5OYD_B GH5 endo-xyloglucanase from Cellvibrio japonicus [Cellvibrio japonicus Ueda107],5OYE_A GH5 endo-xyloglucanase from Cellvibrio japonicus [Cellvibrio japonicus Ueda107],5OYE_B GH5 endo-xyloglucanase from Cellvibrio japonicus [Cellvibrio japonicus Ueda107] |
| 6HA9_A | 5.82e-80 | 133 | 491 | 33 | 393 | Structureof an endo-Xyloglucanase from Cellvibrio japonicus complexed with XXXG(2F)-beta-DNP [Cellvibrio japonicus Ueda107],6HA9_B Structure of an endo-Xyloglucanase from Cellvibrio japonicus complexed with XXXG(2F)-beta-DNP [Cellvibrio japonicus Ueda107],6HAA_A Structure of a covalent complex of endo-Xyloglucanase from Cellvibrio japonicus after reacting with XXXG(2F)-beta-DNP [Cellvibrio japonicus Ueda107],6HAA_B Structure of a covalent complex of endo-Xyloglucanase from Cellvibrio japonicus after reacting with XXXG(2F)-beta-DNP [Cellvibrio japonicus Ueda107] |
| 6WQY_A | 8.54e-72 | 142 | 479 | 21 | 368 | ChainA, Cellulase [Phocaeicola salanitronis DSM 18170] |
| 2JEP_A | 1.25e-65 | 145 | 490 | 35 | 393 | Nativefamily 5 xyloglucanase from Paenibacillus pabuli [Paenibacillus pabuli],2JEP_B Native family 5 xyloglucanase from Paenibacillus pabuli [Paenibacillus pabuli],2JEQ_A Family 5 xyloglucanase from Paenibacillus pabuli in complex with ligand [Paenibacillus pabuli] |
| 3ZMR_A | 7.51e-63 | 144 | 490 | 114 | 465 | Bacteroidesovatus GH5 xyloglucanase in complex with a XXXG heptasaccharide [Bacteroides ovatus],3ZMR_B Bacteroides ovatus GH5 xyloglucanase in complex with a XXXG heptasaccharide [Bacteroides ovatus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O08342 | 1.10e-64 | 144 | 490 | 39 | 398 | Endoglucanase A OS=Paenibacillus barcinonensis OX=198119 GN=celA PE=1 SV=1 |
| A7LXT7 | 7.64e-62 | 144 | 490 | 149 | 500 | Xyloglucan-specific endo-beta-1,4-glucanase BoGH5A OS=Bacteroides ovatus (strain ATCC 8483 / DSM 1896 / JCM 5824 / BCRC 10623 / CCUG 4943 / NCTC 11153) OX=411476 GN=BACOVA_02653 PE=1 SV=1 |
| P28623 | 4.01e-60 | 145 | 475 | 43 | 356 | Endoglucanase D OS=Clostridium cellulovorans (strain ATCC 35296 / DSM 3052 / OCM 3 / 743B) OX=573061 GN=engD PE=1 SV=2 |
| P23660 | 8.11e-60 | 134 | 474 | 15 | 346 | Endoglucanase A OS=Ruminococcus albus OX=1264 GN=celA PE=1 SV=1 |
| P28621 | 3.00e-58 | 144 | 491 | 41 | 374 | Endoglucanase B OS=Clostridium cellulovorans (strain ATCC 35296 / DSM 3052 / OCM 3 / 743B) OX=573061 GN=engB PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
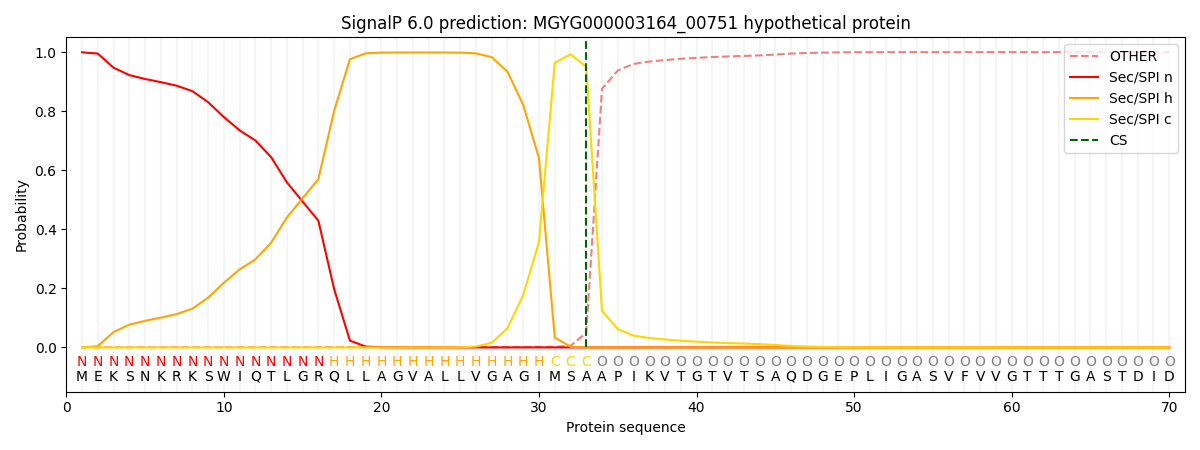
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.001513 | 0.997571 | 0.000297 | 0.000210 | 0.000190 | 0.000182 |