You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003172_00348
You are here: Home > Sequence: MGYG000003172_00348
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | CAG-485 sp900555915 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Muribaculaceae; CAG-485; CAG-485 sp900555915 | |||||||||||
| CAZyme ID | MGYG000003172_00348 | |||||||||||
| CAZy Family | CBM32 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 4055; End: 7420 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM32 | 922 | 1036 | 6.6e-19 | 0.9032258064516129 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00754 | F5_F8_type_C | 4.65e-16 | 922 | 1035 | 6 | 127 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| pfam12708 | Pectate_lyase_3 | 5.72e-15 | 49 | 215 | 3 | 197 | Pectate lyase superfamily protein. This family of proteins possesses a beta helical structure like Pectate lyase. This family is most closely related to glycosyl hydrolase family 28. |
| pfam12708 | Pectate_lyase_3 | 4.42e-04 | 490 | 575 | 5 | 87 | Pectate lyase superfamily protein. This family of proteins possesses a beta helical structure like Pectate lyase. This family is most closely related to glycosyl hydrolase family 28. |
| smart00231 | FA58C | 0.002 | 912 | 1029 | 7 | 130 | Coagulation factor 5/8 C-terminal domain, discoidin domain. Cell surface-attached carbohydrate-binding domain, present in eukaryotes and assumed to have horizontally transferred to eubacterial genomes. |
| COG5434 | Pgu1 | 0.002 | 49 | 117 | 84 | 147 | Polygalacturonase [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUT90069.1 | 3.56e-251 | 29 | 1118 | 27 | 1114 |
| AHW60575.1 | 4.78e-196 | 42 | 1048 | 10 | 980 |
| AEV98037.1 | 4.85e-186 | 22 | 1037 | 24 | 1004 |
| QJD86468.1 | 4.68e-69 | 27 | 774 | 911 | 1641 |
| QHW29527.1 | 1.51e-55 | 41 | 764 | 1093 | 1870 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
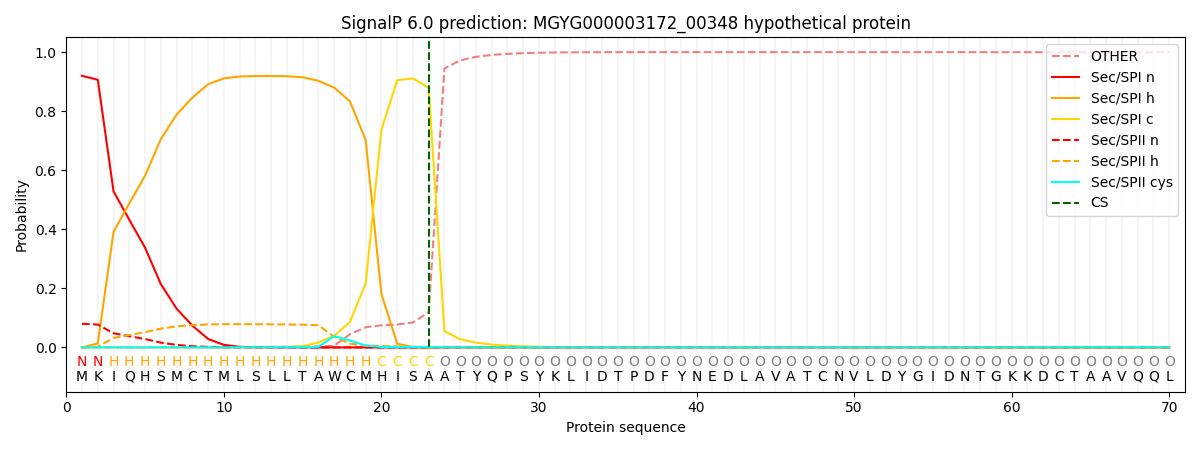
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000805 | 0.914627 | 0.083802 | 0.000284 | 0.000240 | 0.000215 |
