You are browsing environment: HUMAN GUT
MGYG000003172_00463
Basic Information
help
Species
CAG-485 sp900555915
Lineage
Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Muribaculaceae; CAG-485; CAG-485 sp900555915
CAZyme ID
MGYG000003172_00463
CAZy Family
GT83
CAZyme Description
Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase
CAZyme Property
Genome Property
Genome Assembly ID
Genome Size
Genome Type
Country
Continent
MGYG000003172
1933519
MAG
United States
North America
Gene Location
Start: 3844;
End: 5445
Strand: +
No EC number prediction in MGYG000003172_00463.
Family
Start
End
Evalue
family coverage
GT83
11
464
2.3e-60
0.8296296296296296
Cdd ID
Domain
E-Value
qStart
qEnd
sStart
sEnd
Domain Description
COG1807
ArnT
1.08e-20
13
446
11
436
4-amino-4-deoxy-L-arabinose transferase or related glycosyltransferase of PMT family [Cell wall/membrane/envelope biogenesis].
more
Hit ID
E-Value
Query Start
Query End
Hit Start
Hit End
Description
5EZM_A
5.33e-09
8
428
28
446
CrystalStructure of ArnT from Cupriavidus metallidurans in the apo state [Cupriavidus metallidurans CH34],5F15_A Crystal Structure of ArnT from Cupriavidus metallidurans bound to Undecaprenyl phosphate [Cupriavidus metallidurans CH34]
more
This protein is predicted as OTHER
Other
SP_Sec_SPI
LIPO_Sec_SPII
TAT_Tat_SPI
TATLIP_Sec_SPII
PILIN_Sec_SPIII
0.999987
0.000029
0.000002
0.000000
0.000000
0.000015
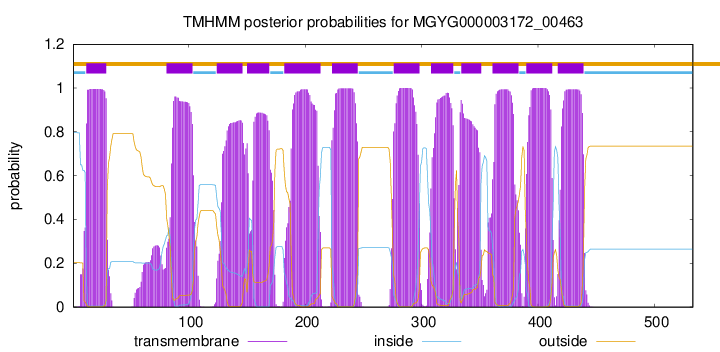
start
end
12
29
81
103
124
146
150
169
182
213
223
245
276
298
308
327
334
351
361
383
390
412
417
439