You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003187_01076
You are here: Home > Sequence: MGYG000003187_01076
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Leclercia adecarboxylata_C | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Proteobacteria; Gammaproteobacteria; Enterobacterales; Enterobacteriaceae; Leclercia; Leclercia adecarboxylata_C | |||||||||||
| CAZyme ID | MGYG000003187_01076 | |||||||||||
| CAZy Family | AA10 | |||||||||||
| CAZyme Description | GlcNAc-binding protein A | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 91495; End: 92955 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA10 | 22 | 188 | 2.7e-58 | 0.9943820224719101 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PRK13211 | PRK13211 | 0.0 | 1 | 486 | 3 | 478 | N-acetylglucosamine-binding protein GbpA. |
| COG3397 | COG3397 | 4.08e-86 | 7 | 300 | 15 | 308 | Predicted carbohydrate-binding protein, contains CBM5 and CBM33 domains [General function prediction only]. |
| cd21177 | LPMO_AA10 | 2.08e-77 | 22 | 188 | 1 | 180 | lytic polysaccharide monooxygenase (LPMO) auxiliary activity family 10 (AA10). AA10 proteins are copper-dependent lytic polysaccharide monooxygenases (LPMOs), which may act on chitin or cellulose. The family used to be called CBM33. Activities in this family include lytic cellulose monooxygenase (C1-hydroxylating) (EC 1.14.99.54), lytic cellulose monooxygenase (C4-dehydrogenating) (EC 1.14.99.56), lytic chitin monooxygenase (EC 1.14.99.53), and lytic xylan monooxygenase/xylan oxidase (glycosidic bond-cleaving) (EC 1.14.99.-). Also included are viral chitin-binding glycoproteins such as fusolin and spheroidin-like proteins. |
| pfam03067 | LPMO_10 | 1.46e-63 | 22 | 187 | 1 | 186 | Lytic polysaccharide mono-oxygenase, cellulose-degrading. This domain is found associated with a wide variety of cellulose binding domains. This is a family of two very closely related proteins that together act as both a C1- and a C4-oxidising lytic polysaccharide mono-oxygenase, degrading cellulose. This domain is also found in baculoviral spheroidins and spindolins, protein of unknown function. |
| pfam18416 | GbpA_2 | 9.53e-27 | 215 | 306 | 8 | 102 | N-acetylglucosamine binding protein domain 2. This domain can be found in N-acetylglucosamine binding protein (GbpA) from Vibrio cholerae, a bacterial pathogen that colonizes the chitinous exoskeleton of zooplankton as well as the human gastrointestinal tract. GbpA binds to GlcNAc oligosaccharides. Structural comparison show that there are distant structural similarities between domain 2 of GbpA and the beta-domain of the flagellin protein p5. It is suggested that this domain interacts with the bacterial surface, and functions to project an alginate binding domain of the protein from the cell surface. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QDK19038.1 | 0.0 | 1 | 486 | 1 | 486 |
| QGU13608.1 | 0.0 | 1 | 486 | 1 | 486 |
| QIK12335.1 | 0.0 | 1 | 486 | 1 | 486 |
| QFH48149.1 | 0.0 | 1 | 486 | 1 | 485 |
| QNP32813.1 | 0.0 | 1 | 486 | 1 | 485 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2XWX_A | 1.56e-85 | 22 | 408 | 1 | 389 | Vibriocholerae colonization factor GbpA crystal structure [Vibrio cholerae],2XWX_B Vibrio cholerae colonization factor GbpA crystal structure [Vibrio cholerae] |
| 2BEM_A | 3.26e-59 | 22 | 188 | 1 | 167 | Crystalstructure of the Serratia marcescens chitin-binding protein CBP21 [Serratia marcescens],2BEM_B Crystal structure of the Serratia marcescens chitin-binding protein CBP21 [Serratia marcescens],2BEM_C Crystal structure of the Serratia marcescens chitin-binding protein CBP21 [Serratia marcescens],2LHS_A Structure of the chitin binding protein 21 (CBP21) [Serratia marcescens] |
| 5WSZ_A | 6.78e-58 | 22 | 191 | 1 | 168 | Crystalstructure of a lytic polysaccharide monooxygenase,BtLPMO10A, from Bacillus thuringiensis [Bacillus thuringiensis serovar kurstaki],5WSZ_B Crystal structure of a lytic polysaccharide monooxygenase,BtLPMO10A, from Bacillus thuringiensis [Bacillus thuringiensis serovar kurstaki],5WSZ_C Crystal structure of a lytic polysaccharide monooxygenase,BtLPMO10A, from Bacillus thuringiensis [Bacillus thuringiensis serovar kurstaki],5WSZ_D Crystal structure of a lytic polysaccharide monooxygenase,BtLPMO10A, from Bacillus thuringiensis [Bacillus thuringiensis serovar kurstaki] |
| 2BEN_A | 7.00e-58 | 22 | 188 | 1 | 167 | Crystalstructure of the Serratia marcescens chitin-binding protein CBP21 Y54A mutant. [Serratia marcescens],2BEN_B Crystal structure of the Serratia marcescens chitin-binding protein CBP21 Y54A mutant. [Serratia marcescens] |
| 6T5Z_A | 8.27e-53 | 22 | 186 | 1 | 168 | Crystalstructure of an AA10 LPMO from Photorhabdus luminescens [Photorhabdus laumondii subsp. laumondii TTO1],6T5Z_B Crystal structure of an AA10 LPMO from Photorhabdus luminescens [Photorhabdus laumondii subsp. laumondii TTO1],6T5Z_C Crystal structure of an AA10 LPMO from Photorhabdus luminescens [Photorhabdus laumondii subsp. laumondii TTO1] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q8EHY2 | 3.06e-131 | 1 | 483 | 7 | 473 | GlcNAc-binding protein A OS=Shewanella oneidensis (strain MR-1) OX=211586 GN=gbpA PE=3 SV=2 |
| Q6LKG5 | 3.60e-119 | 1 | 482 | 3 | 484 | GlcNAc-binding protein A OS=Photobacterium profundum (strain SS9) OX=298386 GN=gbpA PE=3 SV=1 |
| Q5E183 | 8.23e-116 | 4 | 482 | 6 | 488 | GlcNAc-binding protein A OS=Aliivibrio fischeri (strain ATCC 700601 / ES114) OX=312309 GN=gbpA PE=3 SV=1 |
| B5ESR7 | 2.32e-115 | 4 | 482 | 6 | 488 | GlcNAc-binding protein A OS=Aliivibrio fischeri (strain MJ11) OX=388396 GN=gbpA PE=3 SV=1 |
| A7N3J0 | 1.13e-111 | 4 | 482 | 6 | 483 | GlcNAc-binding protein A OS=Vibrio campbellii (strain ATCC BAA-1116 / BB120) OX=1224742 GN=gbpA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
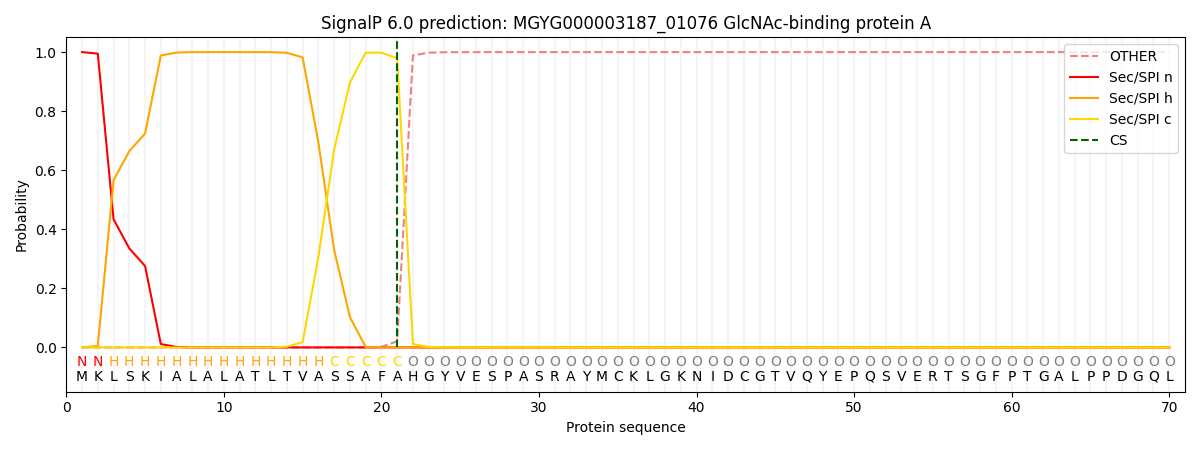
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000276 | 0.999031 | 0.000173 | 0.000174 | 0.000162 | 0.000151 |
