You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003192_01107
You are here: Home > Sequence: MGYG000003192_01107
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
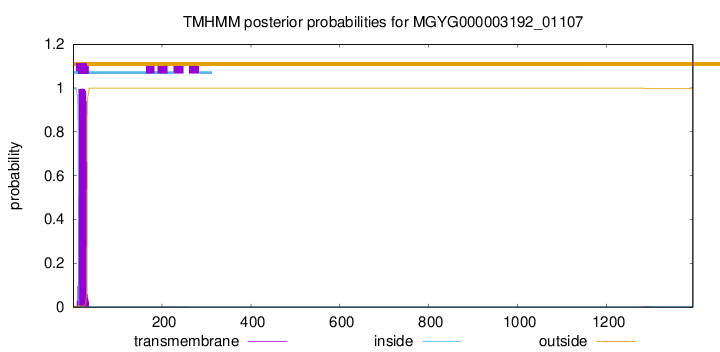
TMHMM annotations
Basic Information help
| Species | UBA5416 sp900539175 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; UBA5416; UBA5416 sp900539175 | |||||||||||
| CAZyme ID | MGYG000003192_01107 | |||||||||||
| CAZy Family | CBM13 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 8071; End: 12255 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam13385 | Laminin_G_3 | 8.39e-18 | 596 | 762 | 1 | 151 | Concanavalin A-like lectin/glucanases superfamily. This domain belongs to the Concanavalin A-like lectin/glucanases superfamily. |
| pfam13385 | Laminin_G_3 | 4.38e-17 | 826 | 978 | 10 | 151 | Concanavalin A-like lectin/glucanases superfamily. This domain belongs to the Concanavalin A-like lectin/glucanases superfamily. |
| pfam13385 | Laminin_G_3 | 5.74e-15 | 1117 | 1190 | 81 | 151 | Concanavalin A-like lectin/glucanases superfamily. This domain belongs to the Concanavalin A-like lectin/glucanases superfamily. |
| pfam05426 | Alginate_lyase | 2.72e-14 | 113 | 314 | 60 | 267 | Alginate lyase. This family contains several bacterial alginate lyase proteins. Alginate is a family of 1-4-linked copolymers of beta -D-mannuronic acid (M) and alpha -L-guluronic acid (G). It is produced by brown algae and by some bacteria belonging to the genera Azotobacter and Pseudomonas. Alginate lyases catalyze the depolymerization of alginates by beta -elimination, generating a molecule containing 4-deoxy-L-erythro-hex-4-enepyranosyluronate at the nonreducing end. This family adopts an all alpha fold. |
| pfam14200 | RicinB_lectin_2 | 1.61e-11 | 456 | 540 | 5 | 89 | Ricin-type beta-trefoil lectin domain-like. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QDA29257.1 | 2.05e-143 | 44 | 553 | 35 | 554 |
| AHM66085.1 | 8.04e-142 | 44 | 553 | 35 | 554 |
| AIY11542.1 | 8.04e-142 | 44 | 553 | 35 | 554 |
| AZH29500.1 | 5.94e-141 | 44 | 553 | 35 | 554 |
| QPK60353.1 | 1.61e-140 | 44 | 553 | 35 | 554 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P19424 | 8.95e-17 | 1207 | 1384 | 40 | 210 | Endoglucanase OS=Bacillus sp. (strain KSM-635) OX=1415 PE=1 SV=1 |
| C6CRV0 | 2.16e-11 | 1218 | 1386 | 1291 | 1457 | Endo-1,4-beta-xylanase A OS=Paenibacillus sp. (strain JDR-2) OX=324057 GN=xynA1 PE=1 SV=1 |
| Q06853 | 2.84e-07 | 1242 | 1386 | 492 | 636 | Cell surface glycoprotein 2 OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=Cthe_3079 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
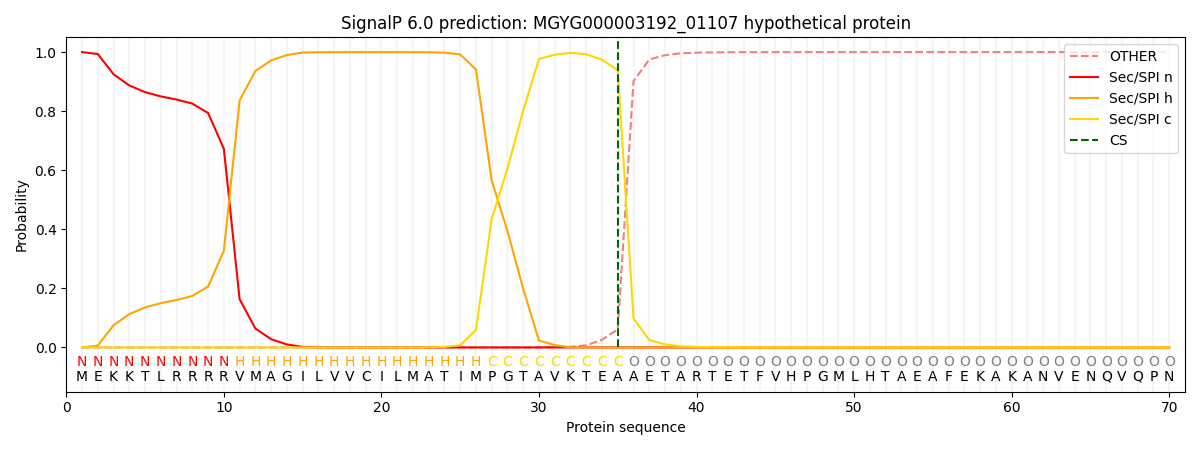
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000282 | 0.998939 | 0.000194 | 0.000236 | 0.000172 | 0.000139 |