You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003192_02009
You are here: Home > Sequence: MGYG000003192_02009
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | UBA5416 sp900539175 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; UBA5416; UBA5416 sp900539175 | |||||||||||
| CAZyme ID | MGYG000003192_02009 | |||||||||||
| CAZy Family | CE1 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 17998; End: 25128 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE1 | 1314 | 1512 | 9.5e-26 | 0.8898678414096917 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG4099 | COG4099 | 5.55e-41 | 1205 | 1532 | 74 | 386 | Predicted peptidase [General function prediction only]. |
| pfam03629 | SASA | 2.00e-25 | 1833 | 2064 | 6 | 223 | Carbohydrate esterase, sialic acid-specific acetylesterase. The catalytic triad of this esterase enzyme comprises residues Ser127, His403 and Asp391 in UniProtKB:P70665. |
| cd02850 | E_set_Cellulase_N | 8.64e-17 | 349 | 424 | 1 | 86 | N-terminal Early set domain associated with the catalytic domain of cellulase. E or "early" set domains are associated with the catalytic domain of cellulases at the N-terminal end. Cellulases are O-glycosyl hydrolases (GHs) that hydrolyze beta 1-4 glucosidic bonds in cellulose. They are usually categorized into either exoglucanases, which sequentially release terminal sugar units from the cellulose chain, or endoglucanases, which also attack the chain internally. The N-terminal domain of cellulase may be related to the immunoglobulin and/or fibronectin type III superfamilies. These domains are associated with different types of catalytic domains at either the N-terminal or C-terminal end and may be involved in homodimeric/tetrameric/dodecameric interactions. Members of this family include members of the alpha amylase family, sialidase, galactose oxidase, cellulase, cellulose, hyaluronate lyase, chitobiase, and chitinase, among others. |
| COG1506 | DAP2 | 9.04e-13 | 1316 | 1516 | 378 | 599 | Dipeptidyl aminopeptidase/acylaminoacyl peptidase [Amino acid transport and metabolism]. |
| COG0400 | YpfH | 9.67e-12 | 1333 | 1508 | 19 | 186 | Predicted esterase [General function prediction only]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BCG58036.1 | 5.12e-195 | 194 | 992 | 216 | 996 |
| SDT49439.1 | 2.94e-92 | 334 | 978 | 211 | 870 |
| BCI61582.1 | 1.40e-52 | 1294 | 1532 | 804 | 1042 |
| ARE59829.1 | 4.57e-38 | 2189 | 2373 | 778 | 962 |
| QQR06511.1 | 4.57e-38 | 2189 | 2373 | 778 | 962 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3DOH_A | 1.16e-45 | 1298 | 1533 | 141 | 380 | CrystalStructure of a Thermostable Esterase [Thermotoga maritima],3DOH_B Crystal Structure of a Thermostable Esterase [Thermotoga maritima],3DOI_A Crystal Structure of a Thermostable Esterase complex with paraoxon [Thermotoga maritima],3DOI_B Crystal Structure of a Thermostable Esterase complex with paraoxon [Thermotoga maritima] |
| 3WYD_A | 2.33e-28 | 1316 | 1524 | 21 | 211 | C-terminalesterase domain of LC-Est1 [uncultured organism],3WYD_B C-terminal esterase domain of LC-Est1 [uncultured organism] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| C6CRV0 | 2.89e-16 | 2196 | 2370 | 1283 | 1457 | Endo-1,4-beta-xylanase A OS=Paenibacillus sp. (strain JDR-2) OX=324057 GN=xynA1 PE=1 SV=1 |
| P19424 | 1.03e-14 | 2195 | 2368 | 40 | 210 | Endoglucanase OS=Bacillus sp. (strain KSM-635) OX=1415 PE=1 SV=1 |
| P38536 | 3.85e-12 | 2220 | 2374 | 1705 | 1857 | Amylopullulanase OS=Thermoanaerobacterium thermosulfurigenes OX=33950 GN=amyB PE=3 SV=2 |
| P38535 | 5.62e-12 | 2220 | 2374 | 931 | 1083 | Exoglucanase XynX OS=Acetivibrio thermocellus OX=1515 GN=xynX PE=3 SV=1 |
| Q06852 | 4.89e-08 | 2211 | 2374 | 2097 | 2266 | Cell surface glycoprotein 1 OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=olpB PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
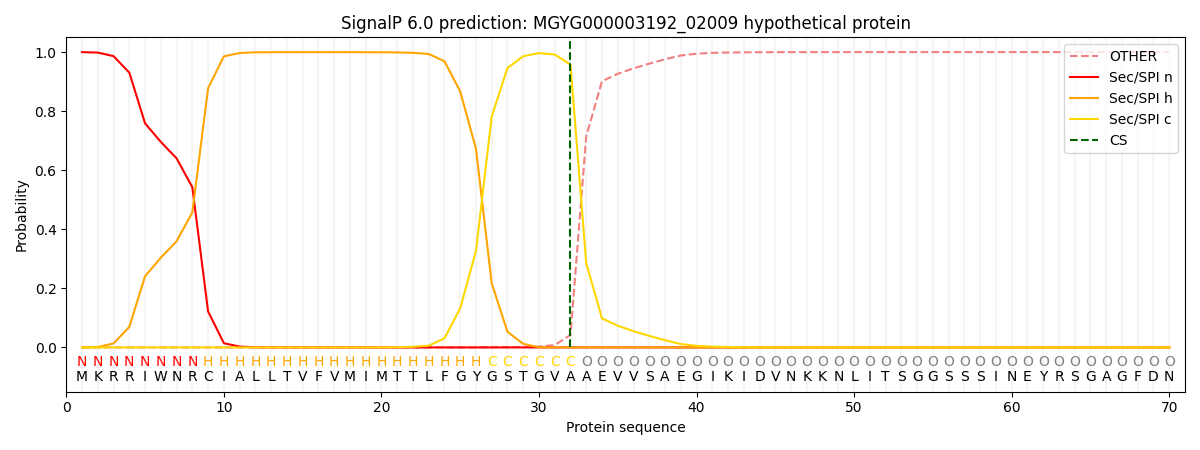
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000505 | 0.998612 | 0.000365 | 0.000180 | 0.000153 | 0.000144 |

