You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003194_01110
You are here: Home > Sequence: MGYG000003194_01110
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
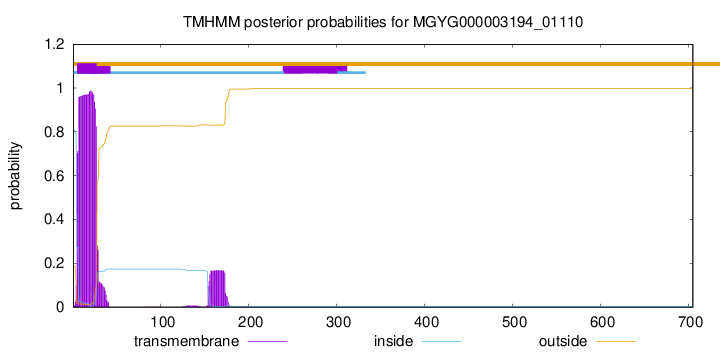
TMHMM annotations
Basic Information help
| Species | Paramuribaculum sp900546365 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Muribaculaceae; Paramuribaculum; Paramuribaculum sp900546365 | |||||||||||
| CAZyme ID | MGYG000003194_01110 | |||||||||||
| CAZy Family | GH89 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 16423; End: 18540 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH89 | 74 | 699 | 1.3e-204 | 0.9532428355957768 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam05089 | NAGLU | 8.76e-154 | 128 | 452 | 1 | 333 | Alpha-N-acetylglucosaminidase (NAGLU) tim-barrel domain. Alpha-N-acetylglucosaminidase, a lysosomal enzyme required for the stepwise degradation of heparan sulfate. Mutations on the alpha-N-acetylglucosaminidase (NAGLU) gene can lead to Mucopolysaccharidosis type IIIB (MPS IIIB; or Sanfilippo syndrome type B) characterized by neurological dysfunction but relatively mild somatic manifestations. The structure shows that the enzyme is composed of three domains. This central domain has a tim barrel fold. |
| pfam12972 | NAGLU_C | 6.61e-79 | 459 | 703 | 1 | 242 | Alpha-N-acetylglucosaminidase (NAGLU) C-terminal domain. Alpha-N-acetylglucosaminidase, a lysosomal enzyme required for the stepwise degradation of heparan sulfate. Mutations on the alpha-N-acetylglucosaminidase (NAGLU) gene can lead to Mucopolysaccharidosis type IIIB (MPS IIIB; or Sanfilippo syndrome type B) characterized by neurological dysfunction but relatively mild somatic manifestations. The structure shows that the enzyme is composed of three domains. This C-terminal domain has an all alpha helical fold. |
| pfam12971 | NAGLU_N | 7.22e-23 | 40 | 113 | 6 | 81 | Alpha-N-acetylglucosaminidase (NAGLU) N-terminal domain. Alpha-N-acetylglucosaminidase, a lysosomal enzyme required for the stepwise degradation of heparan sulfate. Mutations on the alpha-N-acetylglucosaminidase (NAGLU) gene can lead to Mucopolysaccharidosis type IIIB (MPS IIIB; or Sanfilippo syndrome type B) characterized by neurological dysfunction but relatively mild somatic manifestations. The structure shows that the enzyme is composed of three domains. This N-terminal domain has an alpha-beta fold. |
| cd19962 | PBP1_ABC_RfuA-like | 0.002 | 232 | 350 | 105 | 226 | periplasmic riboflavin-binding component (RfuA) of ABC transporter (RfuABCD) from Treponema pallidum and its close homologs in other bacteria. This group includes the basic membrane lipoprotein (BMP) family ABC transporter substrate-binding protein RfuA from Treponema pallidum and its close homologs in other bacteria. RfuA is the riboflavin-binding component of ABC transporter (RfuABCD) in spirochetes. The members of this group are highly similar to that of the periplasmic binding domain of basic membrane lipoprotein (BMP), PnrA. The PnrA lipoprotein, also known as Tp0319 or TmpC, represents a novel family of bacterial purine nucleoside receptor encoded within an ATP-binding cassette (ABC) transport system (pnrABCDE). It shows a striking structural similarity to another basic membrane lipoprotein Med which regulates the competence transcription factor gene, comK, in Bacillus subtilis. PnrA-like proteins are likely to have similar nucleoside-binding functions and a similar type 1 periplasmic sugar-binding protein-like fold. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QRO24616.1 | 1.90e-255 | 15 | 696 | 1 | 686 |
| QUT50280.1 | 4.39e-254 | 34 | 696 | 19 | 686 |
| ADV42401.1 | 1.40e-242 | 16 | 705 | 3 | 695 |
| QCQ40346.1 | 5.63e-242 | 20 | 705 | 6 | 697 |
| CBW21233.1 | 5.63e-242 | 20 | 705 | 6 | 697 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2VC9_A | 2.74e-126 | 61 | 703 | 201 | 850 | Family89 Glycoside Hydrolase from Clostridium perfringens in complex with 2-acetamido-1,2-dideoxynojirmycin [Clostridium perfringens],2VCA_A Family 89 glycoside hydrolase from Clostridium perfringens in complex with beta-N-acetyl-D-glucosamine [Clostridium perfringens],2VCB_A Family 89 Glycoside Hydrolase from Clostridium perfringens in complex with PUGNAc [Clostridium perfringens],2VCC_A Family 89 Glycoside Hydrolase from Clostridium perfringens [Clostridium perfringens] |
| 7MFK_A | 3.31e-126 | 61 | 703 | 209 | 858 | ChainA, Alpha-N-acetylglucosaminidase family protein [Clostridium perfringens ATCC 13124],7MFL_A Chain A, Alpha-N-acetylglucosaminidase family protein [Clostridium perfringens ATCC 13124] |
| 4A4A_A | 3.57e-125 | 61 | 703 | 224 | 873 | CpGH89(E483Q, E601Q), from Clostridium perfringens, in complex with its substrate GlcNAc-alpha-1,4-galactose [Clostridium perfringens] |
| 4XWH_A | 1.65e-100 | 62 | 682 | 40 | 662 | Crystalstructure of the human N-acetyl-alpha-glucosaminidase [Homo sapiens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P54802 | 2.17e-99 | 62 | 682 | 63 | 685 | Alpha-N-acetylglucosaminidase OS=Homo sapiens OX=9606 GN=NAGLU PE=1 SV=2 |
| Q9FNA3 | 4.33e-88 | 70 | 703 | 92 | 776 | Alpha-N-acetylglucosaminidase OS=Arabidopsis thaliana OX=3702 GN=NAGLU PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
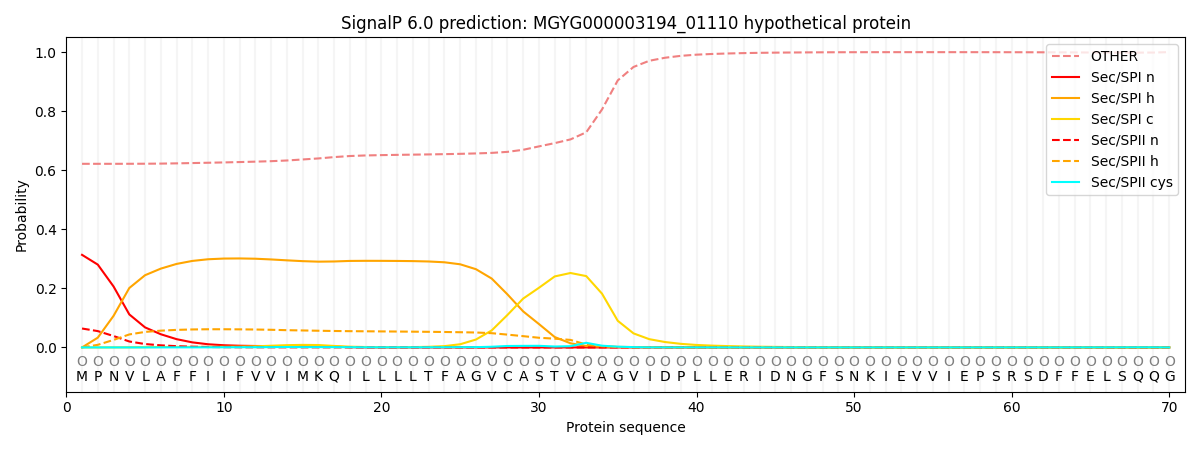
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.632664 | 0.300152 | 0.065501 | 0.000333 | 0.000228 | 0.001126 |