You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003200_01166
You are here: Home > Sequence: MGYG000003200_01166
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | CAG-873 sp900759845 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Muribaculaceae; CAG-873; CAG-873 sp900759845 | |||||||||||
| CAZyme ID | MGYG000003200_01166 | |||||||||||
| CAZy Family | PL9 | |||||||||||
| CAZyme Description | Pectate lyase L | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 2384; End: 3838 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam13229 | Beta_helix | 2.27e-10 | 137 | 318 | 2 | 157 | Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
| pfam13229 | Beta_helix | 1.24e-05 | 151 | 323 | 3 | 138 | Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
| pfam07602 | DUF1565 | 0.001 | 28 | 63 | 3 | 38 | Protein of unknown function (DUF1565). These proteins share a region of homology in their N termini, and are found in several phylogenetically diverse bacteria and in the archaeon Methanosarcina acetivorans. Some of these proteins also contain characterized domains such as pfam00395 and pfam03422. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QQA07680.1 | 7.60e-137 | 1 | 431 | 1 | 434 |
| QMW86526.1 | 1.08e-136 | 1 | 431 | 1 | 434 |
| AAO79288.1 | 1.08e-136 | 1 | 431 | 1 | 434 |
| QIU94582.1 | 2.25e-136 | 1 | 437 | 1 | 443 |
| QUT42080.1 | 4.33e-136 | 1 | 431 | 1 | 434 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5OLQ_A | 7.89e-99 | 23 | 420 | 6 | 419 | Rhamnogalacturonanlyase [Bacteroides thetaiotaomicron],5OLQ_B Rhamnogalacturonan lyase [Bacteroides thetaiotaomicron],5OLQ_C Rhamnogalacturonan lyase [Bacteroides thetaiotaomicron],5OLR_A Rhamnogalacturonan lyase [Bacteroides thetaiotaomicron],5OLR_B Rhamnogalacturonan lyase [Bacteroides thetaiotaomicron],5OLR_C Rhamnogalacturonan lyase [Bacteroides thetaiotaomicron],5OLS_A Rhamnogalacturonan lyase [Bacteroides thetaiotaomicron] |
| 1RU4_A | 6.13e-45 | 23 | 420 | 18 | 375 | ChainA, Pectate lyase [Dickeya chrysanthemi] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P0C1A7 | 5.64e-44 | 23 | 420 | 43 | 400 | Pectate lyase L OS=Dickeya dadantii (strain 3937) OX=198628 GN=pelL PE=1 SV=1 |
| P0C1A6 | 1.49e-42 | 23 | 420 | 43 | 400 | Pectate lyase L OS=Dickeya chrysanthemi OX=556 GN=pelL PE=3 SV=1 |
| P94576 | 8.94e-09 | 20 | 141 | 34 | 172 | Uncharacterized protein YwoF OS=Bacillus subtilis (strain 168) OX=224308 GN=ywoF PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
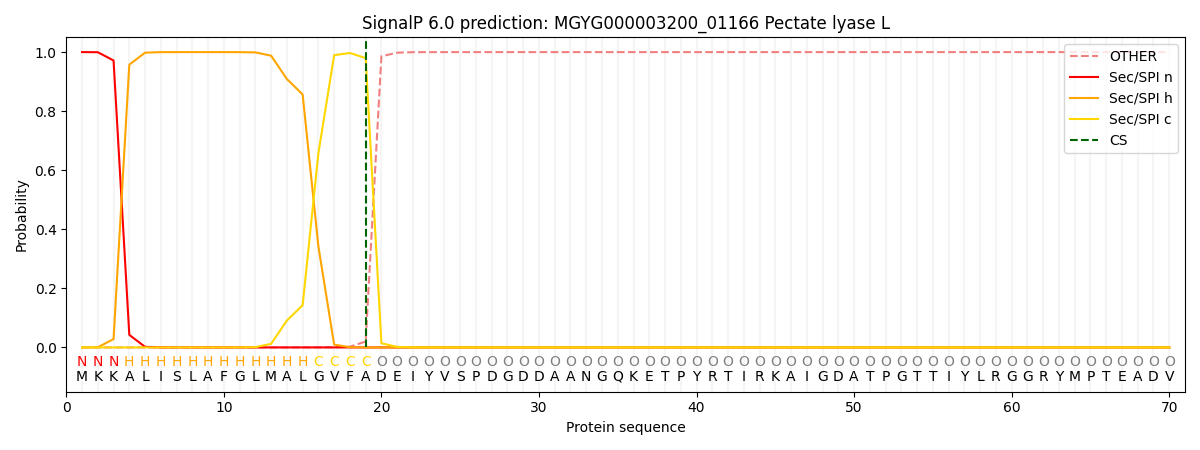
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000308 | 0.998863 | 0.000252 | 0.000184 | 0.000184 | 0.000167 |
