You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003201_00107
You are here: Home > Sequence: MGYG000003201_00107
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | CAG-873 sp900759825 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Muribaculaceae; CAG-873; CAG-873 sp900759825 | |||||||||||
| CAZyme ID | MGYG000003201_00107 | |||||||||||
| CAZy Family | PL29 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 931; End: 2514 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL29 | 86 | 391 | 6.6e-84 | 0.9966777408637874 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam16315 | DUF4955 | 2.34e-52 | 394 | 526 | 1 | 139 | Domain of unknown function (DUF4955). This family consists of uncharacterized proteins around 850 residues in length and is mainly found in various Bacteroides species. The function of this protein is unknown. |
| pfam12708 | Pectate_lyase_3 | 1.10e-10 | 91 | 160 | 6 | 68 | Pectate lyase superfamily protein. This family of proteins possesses a beta helical structure like Pectate lyase. This family is most closely related to glycosyl hydrolase family 28. |
| COG5434 | Pgu1 | 3.63e-05 | 86 | 129 | 82 | 124 | Polygalacturonase [Carbohydrate transport and metabolism]. |
| pfam13229 | Beta_helix | 0.001 | 340 | 434 | 36 | 131 | Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QIL38354.1 | 4.90e-104 | 52 | 526 | 25 | 506 |
| ADE56073.1 | 1.87e-103 | 51 | 527 | 29 | 512 |
| QZT38145.1 | 7.12e-102 | 51 | 526 | 26 | 507 |
| QDU32698.1 | 2.72e-99 | 49 | 526 | 32 | 515 |
| QDM10828.1 | 1.53e-98 | 50 | 526 | 320 | 822 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2PYG_A | 7.44e-07 | 87 | 162 | 3 | 77 | Azotobactervinelandii Mannuronan C-5 epimerase AlgE4 A-module [Azotobacter vinelandii],2PYG_B Azotobacter vinelandii Mannuronan C-5 epimerase AlgE4 A-module [Azotobacter vinelandii],2PYH_A Azotobacter vinelandii Mannuronan C-5 epimerase AlgE4 A-module complexed with mannuronan trisaccharide [Azotobacter vinelandii],2PYH_B Azotobacter vinelandii Mannuronan C-5 epimerase AlgE4 A-module complexed with mannuronan trisaccharide [Azotobacter vinelandii] |
| 5ZRU_A | 3.11e-06 | 89 | 192 | 189 | 295 | ChainA, Alpha-1,3-glucanase [Niallia circulans],5ZRU_C Chain C, Alpha-1,3-glucanase [Niallia circulans] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q44493 | 5.48e-06 | 87 | 162 | 3 | 77 | Mannuronan C5-epimerase AlgE4 OS=Azotobacter vinelandii OX=354 GN=algE4 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
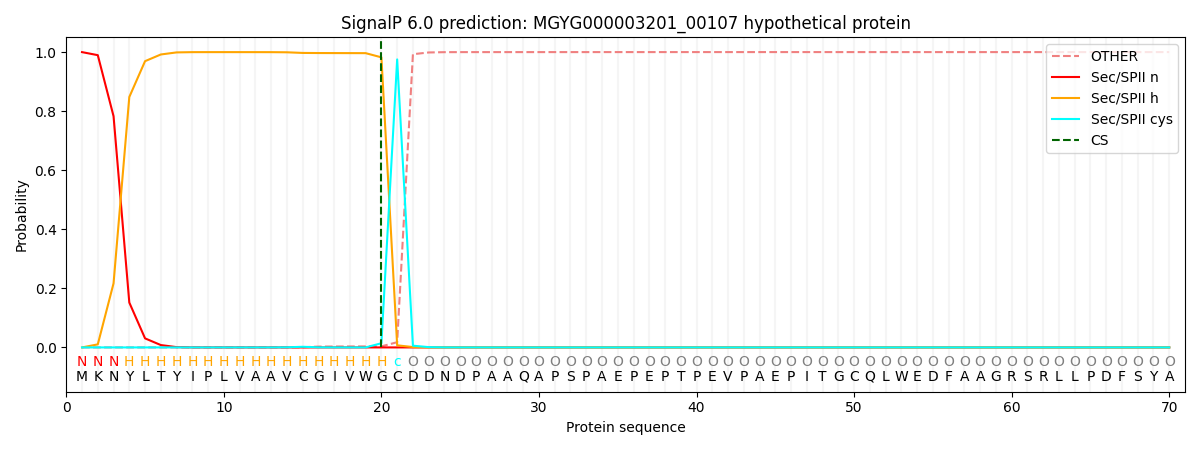
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000091 | 0.999953 | 0.000000 | 0.000000 | 0.000000 |
